...
First you need to prepare a germplasm list with test and check entries to be planted in the trial. We are going to use the list of imported germplam we created in the Import Germplasm Tutorial. Probably called <your initials>GI21– CGMGI21 for me. Use LISTS>GermplasmLists>Browse to view it.

Open the Manage Studies application from the STUDIES menu and click on Start a new study. Name the trial <your initials>21PVT, CGM21PVT for me, and fill in the basic details of description and objective and select the study type Trial.
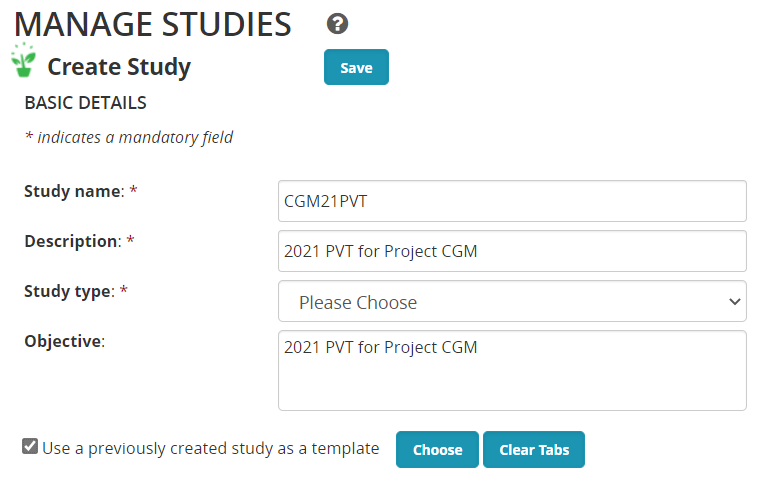
You can add variables to the new study directly from the Ontology pick lists as we did for the crossing block, or you can pick up all the variables from any previously used study. To use this option check the Use a previously created study as a template check box. Then choose the previous study which is smilar to your current study. We dont have a previous trial yet so we will use a nursery:
The variables from the template study are imported into the new study. Change the settings if needed and Save the study in a new folder <your initials> 2021 Trials (CGM 2021 Trials for me).


On the Germplasm and Checks tab, the extra variables for Cross and Seed source are also recovered from the template, so you just need to Browse for the list of planting material. The imported germplasm list you made in the Import Germplasm Tutorial. CGM21GI in my case.
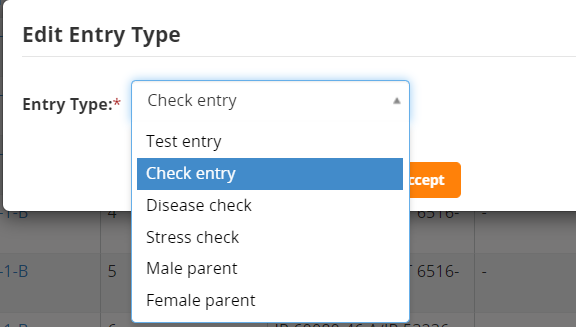
You can set the Check entries on the Germplasm Tab. Click on the entry type for the line(s) to be made checks, (we will make IRRI 132 a check entry), choose Check Entry from the list and click accept.
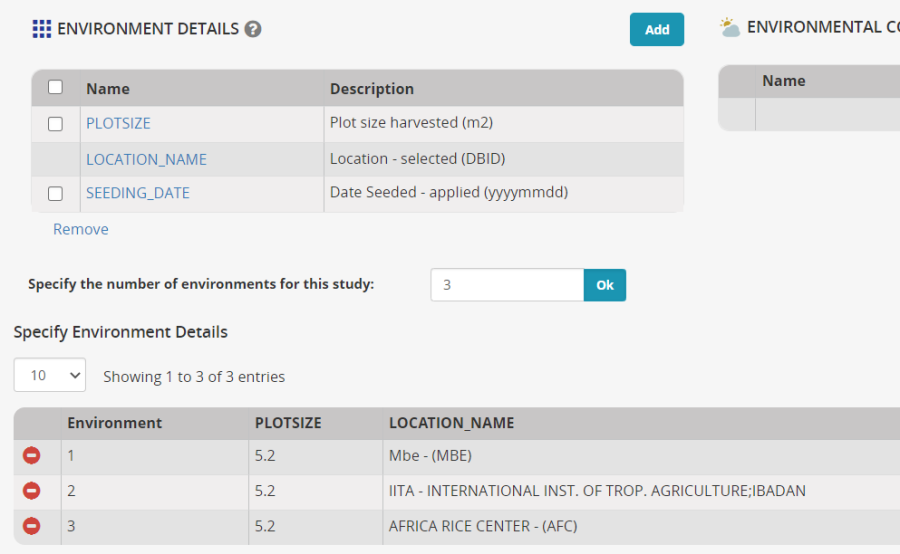
On the Environments tab set the number of environments to 3 and click OK.
Mbe has been inherited from the template for the first environment, and this location is still correct, select IITA-Ibadan and Africa Rice CENTRE as the other two locations. You will have to uncheck Show Favorite Locations since we have not specfied favorite locations. Click Add next to Environmental Details variable, and and search for Seeding date and Plotsize and add them to the Environment Details section. Enter 5.2 m squared for the plot size at each location. You may not know the Seeding date at this time.
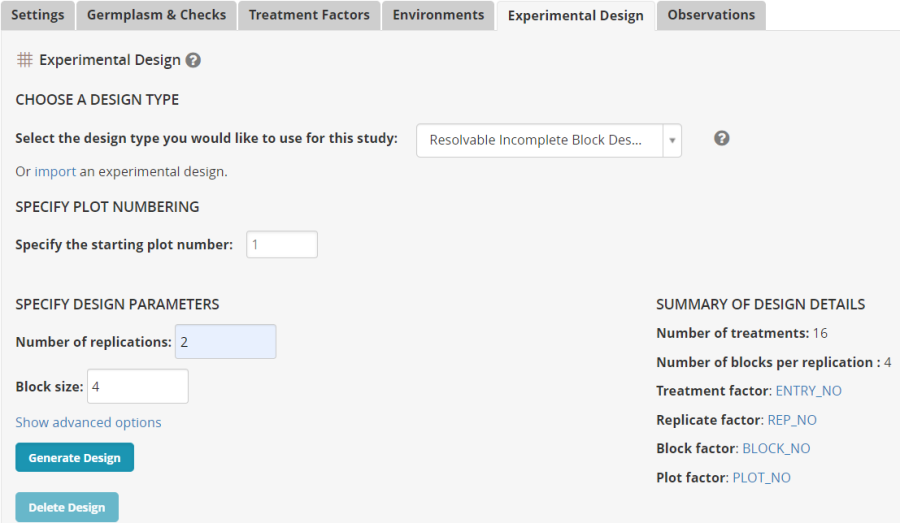
On the Experimental Design tab select Resolveable Resolvable Incomplete Block Design (Alpha lattice), enter 2 for number of replications and 4 for block size then click Generate Design.
Generate the design for all locations:

You will get a success message and now the Observation tab will contain a fieldbook. At the moment the only trait there is the date of 50% flowering inherited from our nursery so we will add some more traits. Click Add next to the traits box. Search for FLW50 and add it to the trial. (If you cant find FLW50 use TFlwS_CM-d).

Notice that trait TFlwS_CM-d has Alias FLW50 and has a computation formula associated so that it can be calculated within BMS.
Add GrYld_wgh_gplot, then GRMOIST, and finally GYTHA which is also a variable which can be calculated in BMS.
The traits box now looks like this:

Prepare seed and planting labels
...










