...
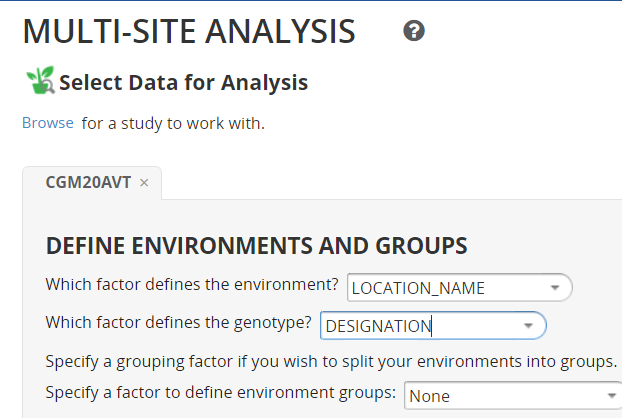
From the STUDIES menu select Multi-Site Analysis and browse to the study for which you have uploaded means and for which you wish to do a multi-site analysis. You need at least three sites for a GxE analysis and four or more is better. As with the single site analysis you are asked to specify the variable defining sites – usually use LOCATION_NAME, and for genotypes – usually DESIGNATION. Next the form asks if your environments are already grouped in some way which would account for significant GxE interactions. Usually we do not know about groupings at the early stage, and mostly do not have enough environments for subsets.
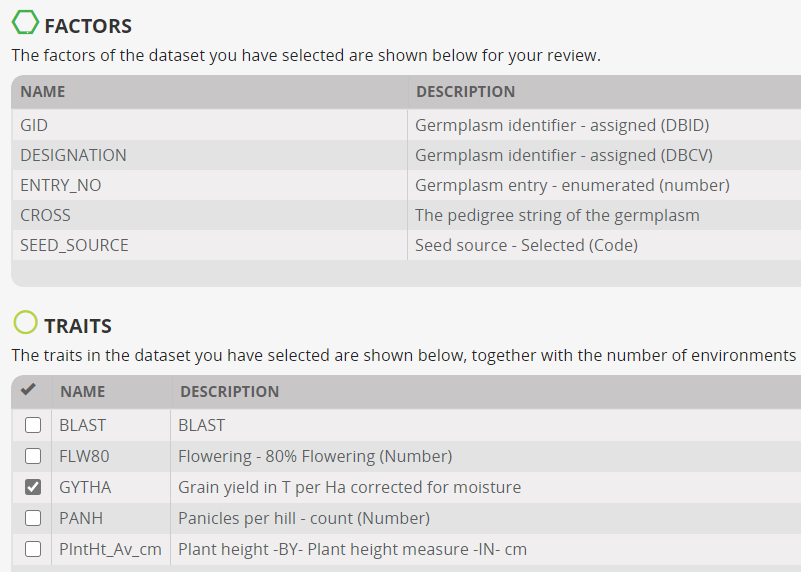
The form also show the variables in the study and asks the user to check the traits for which a GxE analysis is required. We will analysis analyse only GYTHA. Click Next.
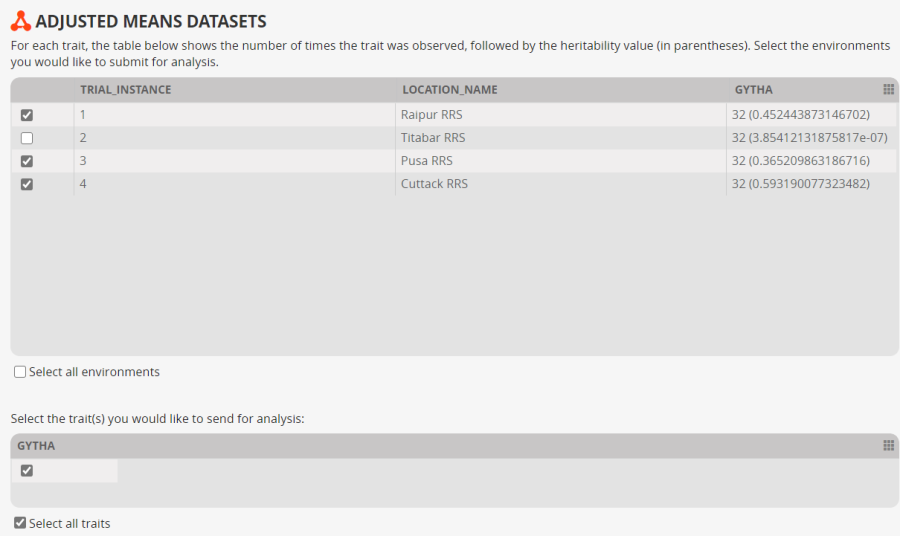
A form displaying a data summary for the locations and the selected traits is presented. This allows you to eliminate environments for which there is insufficient data of for which the heritability is too low, and similarly at the bottom you can eliminate traits. We have already limited out our traits to one, but we notice that Environment 2 has zero heritability. There would not normally be any reason to include an environment in a GxE alanysis analysis which was showing a very small heritability so we will uncheck environment 2 and continue the analysis with just three environments.
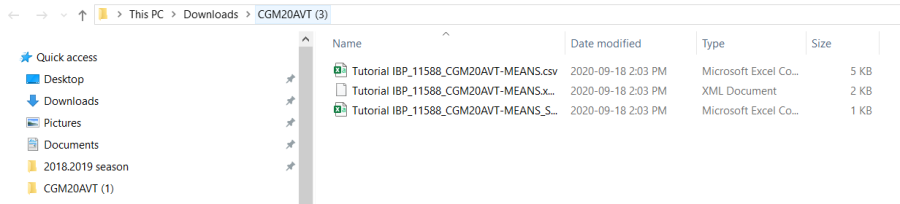
Click Download Input Files at the bottom of the form.
A zip file with the name of the study will be downloaded. You could extract the three files in this zip to a directory for analysis.
...