Making a crossing block
A Crossing Block is a nursery planted with parental material for the purpose of making crosses between lines planted in the nursery. In BMS all activities involving planting material are referred to as studies and so nurseries are one kind of study and we use the Study Manager to manage the information for nurseries.
There are two ways a cross list can be formed. Either by making a planned series of crosses by matching parents from a parent list which is planted in a crossing bloc nursery or by recording crosses made in the field in such a nursery using a crossing template. We will demonstrate both options in this tutorial.
Objectives
At the end of this chapter, the user should be able to:
- Use the Study Manager to create a Nursery, add meta data to the nursery, specify planting material, specify the planting location and create a fieldbook.
- Use the Crossing Manager to make crosses between matched female and male entries in a crossing block.
- Specify a naming convention to give the crosses names in a series.
- Use a crossing template to record crosses made in a crossing block in the field and then load the information into the BMS
Create a Crossing Block in the Study Manager
Click on Manage Studies from the STUDIES menu and then on Start a new Study.
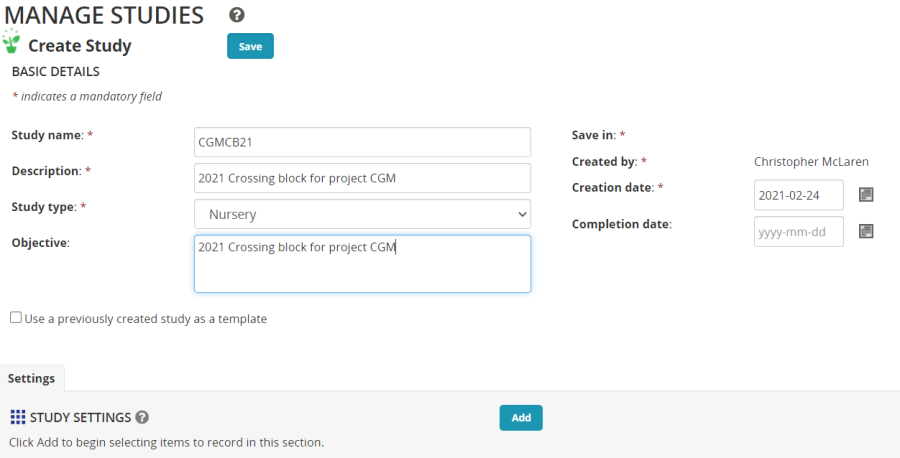
The Create Study form opens. Fill in the basic details with a short Study name (Use your initials in the study name to ensure uniqueness in the class). A description, a study type – Nursery (A nursery is a single location unreplicated trial for population development or germplasm characterization). Give an objective.
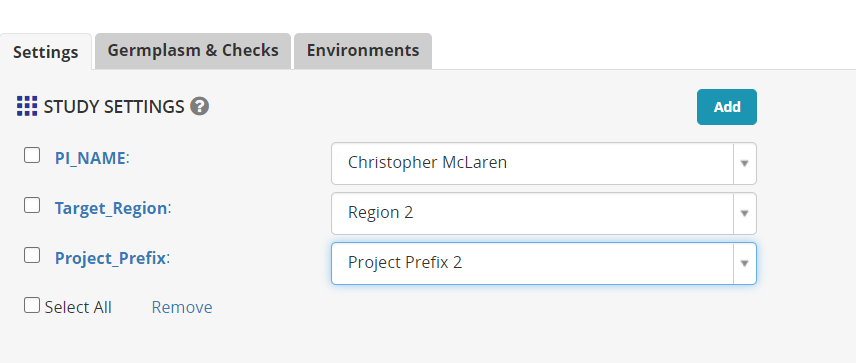
Next click on the Add button next to STUDY SETTINGS. This will open the ontology manager and you may specify variables to be added to the study to record metadata about the study.
In the ontology search box, type PI and add PI Name. Look for region and add Target Region and then look for Project and add Project Prefix. These variables can be customized to your particular programs and identify important objectives of the crossing block.
Now save the nursery definition as created so far by clicking on the Save button on the top left of the Study Manager screen.
If there is no folder for <your initials> 2021 Nurseries, create one with your initials to keep your work separate from other students. (To make a folder, click on the + Add folder symbol, Type the name and click the tick symbol)
The study will be saved and reloaded with a few new tabs.
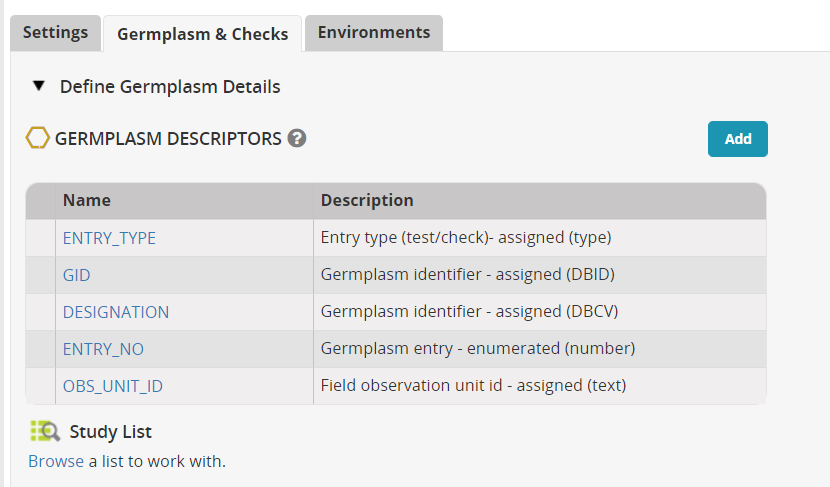
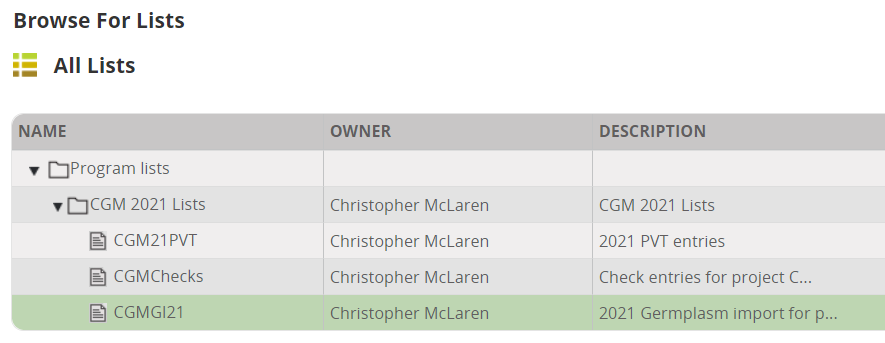
Now click on the tab Germplasm and Checks. Click the Add button next to Germplasm Descriptors.
In the Ontology search box look for 'cross' and add variable Cross to the descriptors, then search for 'source' and add Seed Source to the descriptors.
Now click Browse and navigate through the program lists to your imported list and click Select.
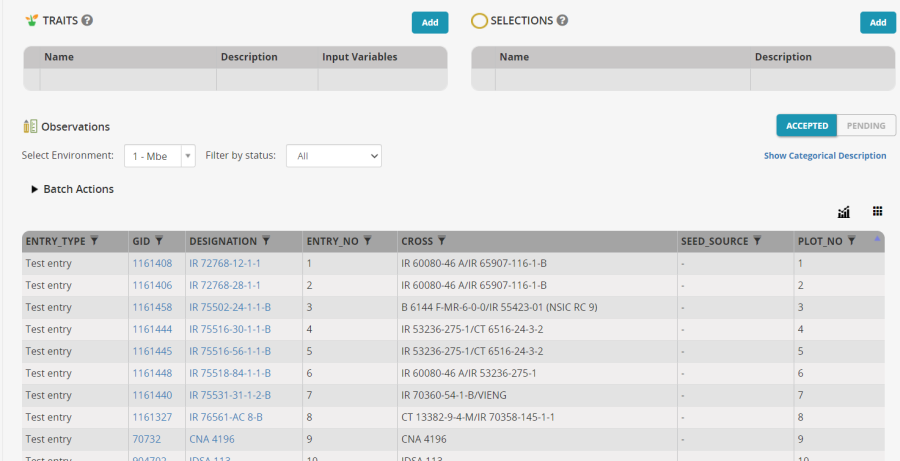
The entries will be imported into the nursery:
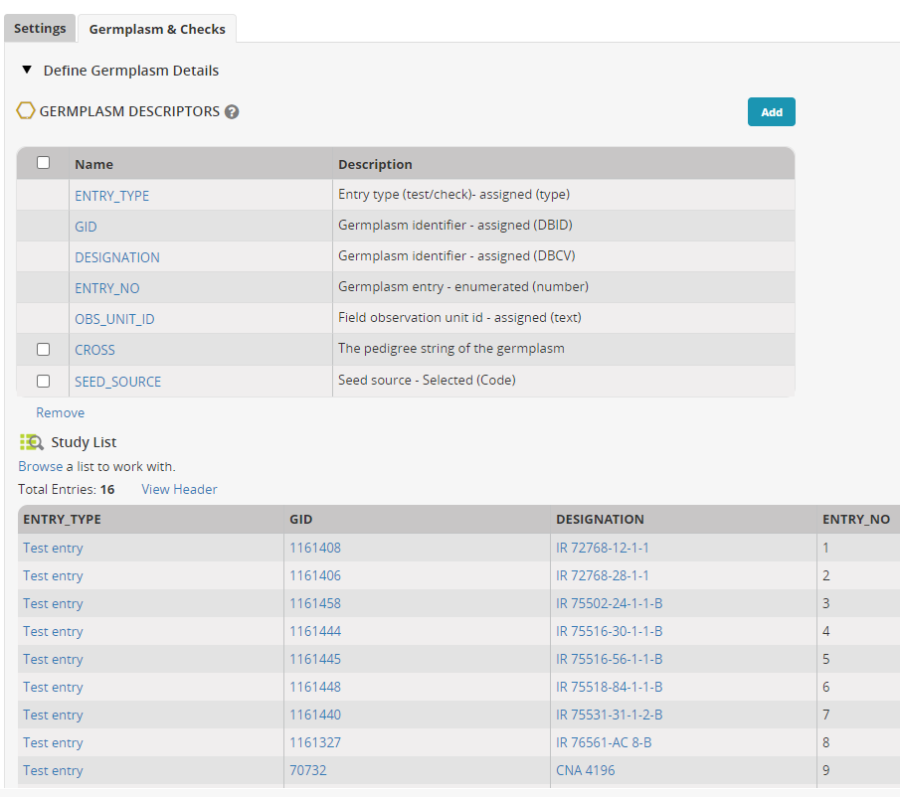
Click on the Environments tab and choose Mbe for the planting location. Click on the Add button next to ENVIRONMENT DETAIL and search for Planting Date in the ontology. You will find SEEDING_DATE – add it to the nursery.
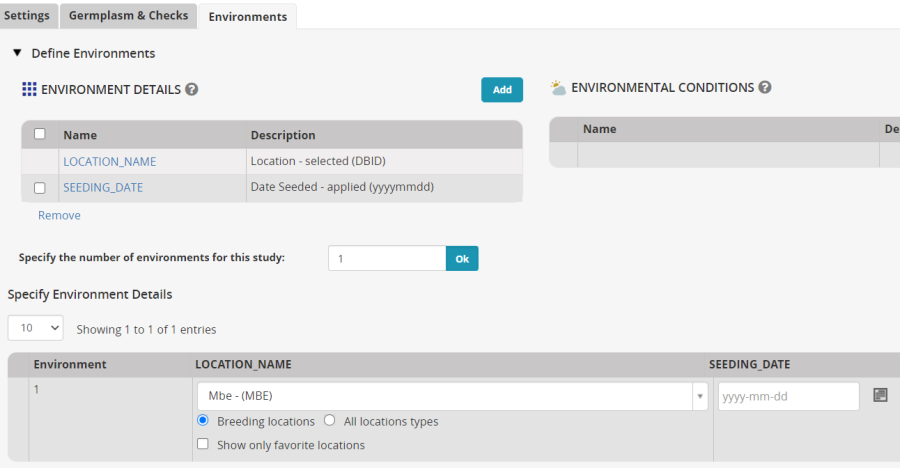
Click on the Experimental Design tab. Select the Design Entry List Order and click Generate Design.
Select the location (we only have the one) and click Generate again. A fieldbook is produced in the observation tab:
It has 16 plots since there is no replication and the entries are planted in the same order as their entry numbers 1 … 16. This is a reasonable lay-out for a crossing block, and indeed for most nurseries. However absolutely any lay-out can be specified by using the Import design function on the Experimental design tab. We will not use this for this exercise.
Use the Crossing Design Tool for Specifying Crosses
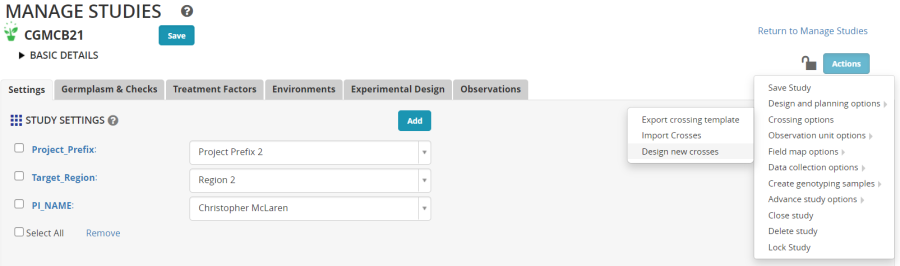
On the Actions Menu, select Crossing Options and then Design new Crosses.
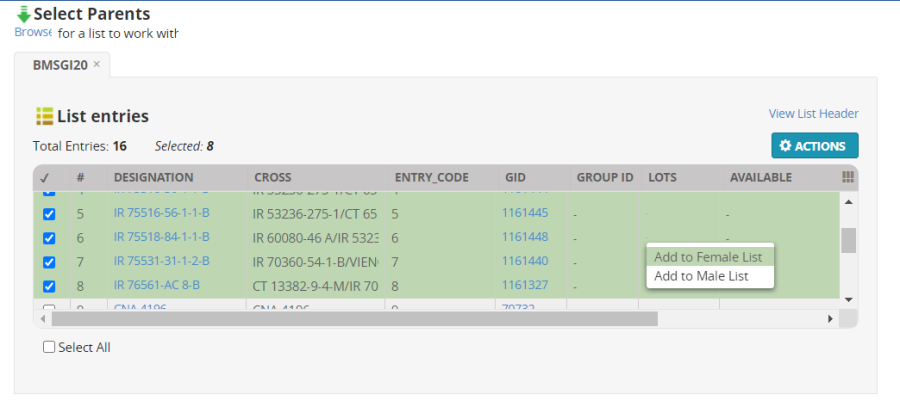
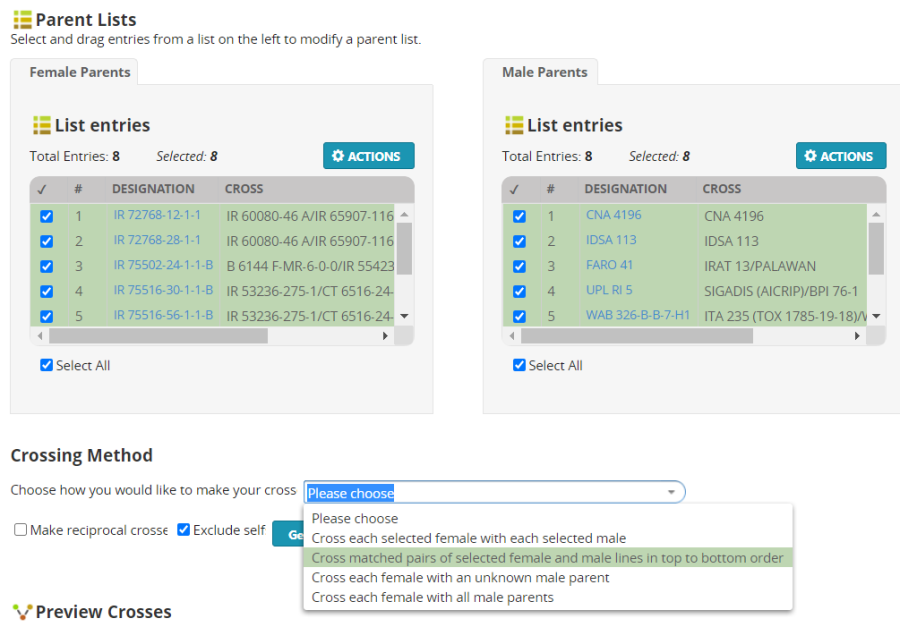
In the Select parents form, highlight the first eight entries (by clicking in the check boxes next to the names) then right click on the green space and select Add to Female List:
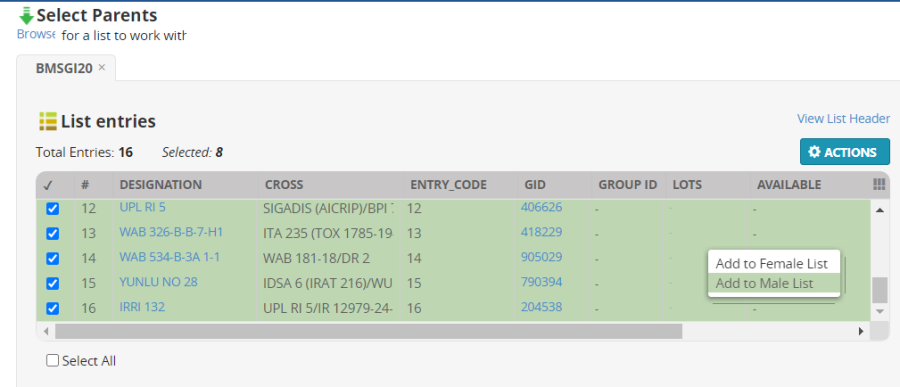
Uncheck the first eight entries and check the last eight entries and select Add to Male List:
The parent lists now look as follows:
There are several ways to combine parents from the female and male lists. For this exercise we are going to choose Cross each selected female with each selected male.
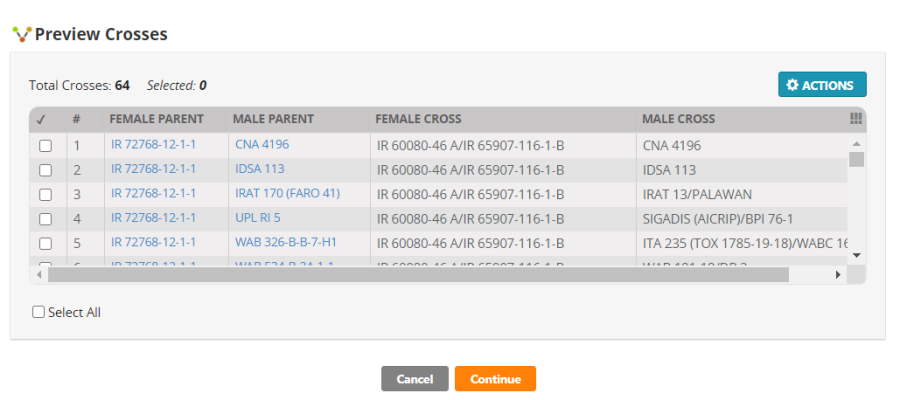
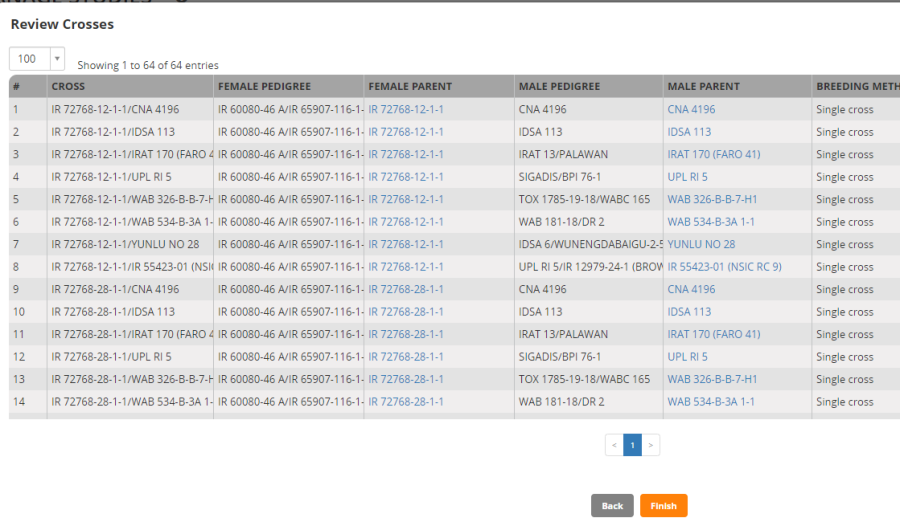
Click Generate Crosses. You will get a preview of 64 crosses:
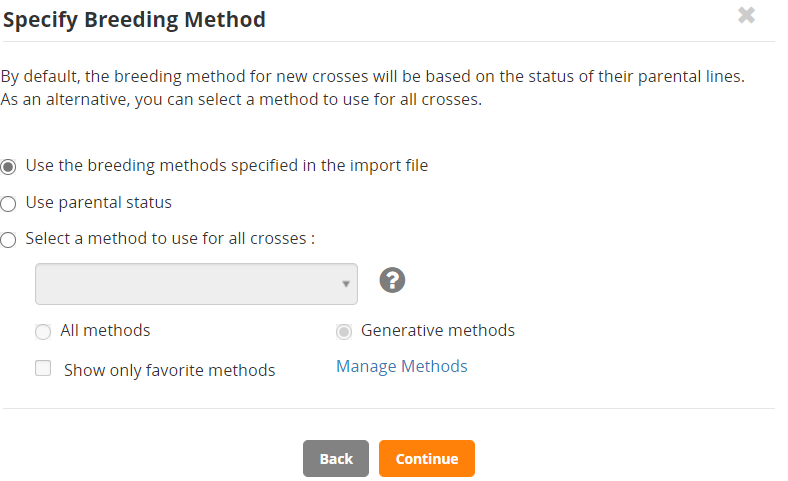
If the crosses are the ones you want to make click Continue. Then you need to specify the method of crossing.
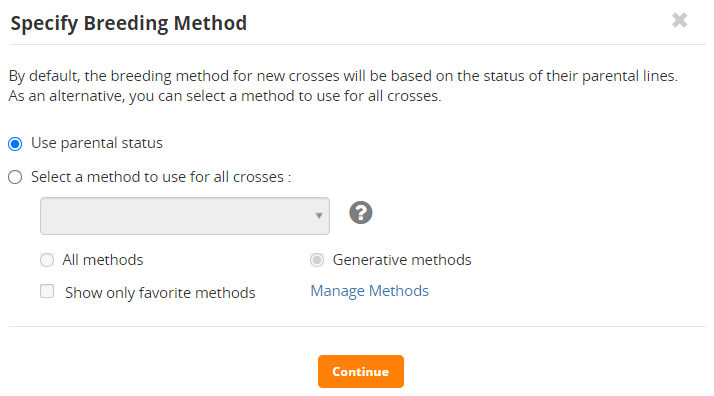
You can select a crossing method for all the crosses from the list at the end of this tutorial by checking the Select a method to use for all crosses radio button and then selecting in the box, or you can allow BMS to work out the type of cross being made by using Use parental status. We will use the parental status option.
Click Continue.
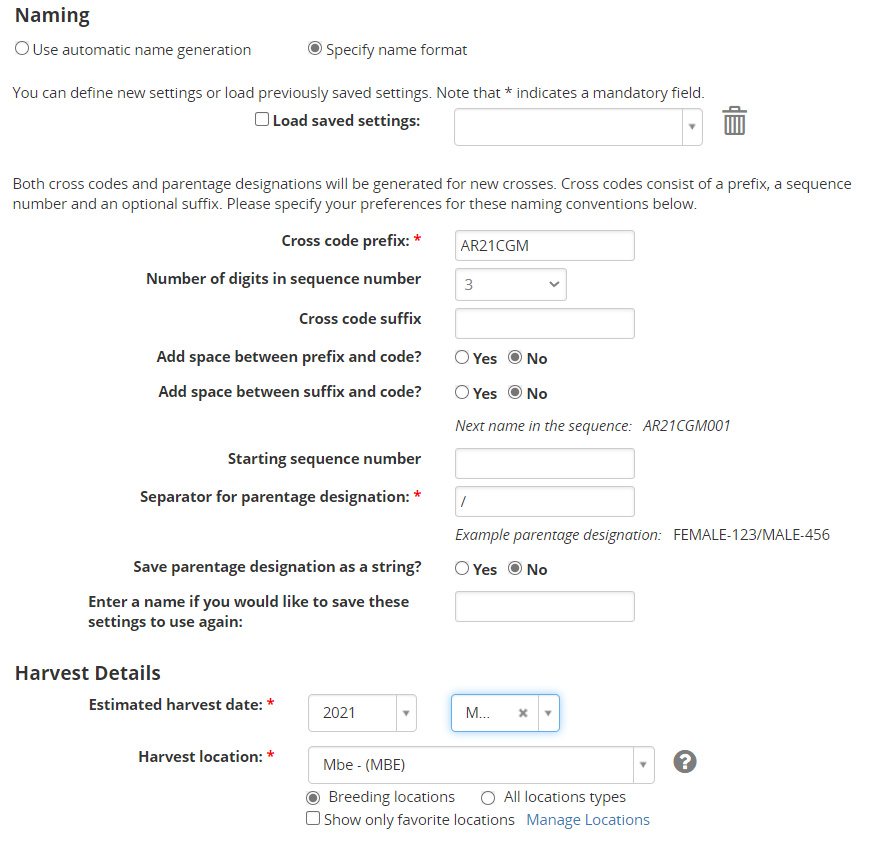
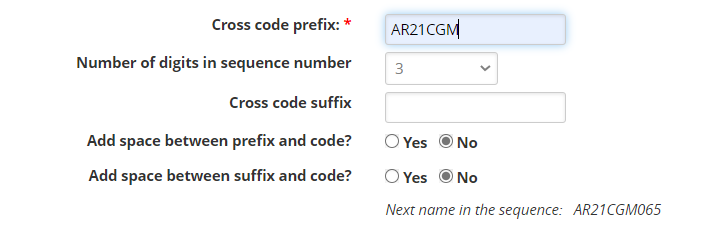
You will be asked to specify Naming and Harvest Details. Click the Specify name format radio button.
- Enter AR21<your initials> as prefix of the Cross Code
- Enter 3 as the number of digits for the sequence code.
- Specify date and location.
Click Continue. You will see a review panel.
Click Finish.
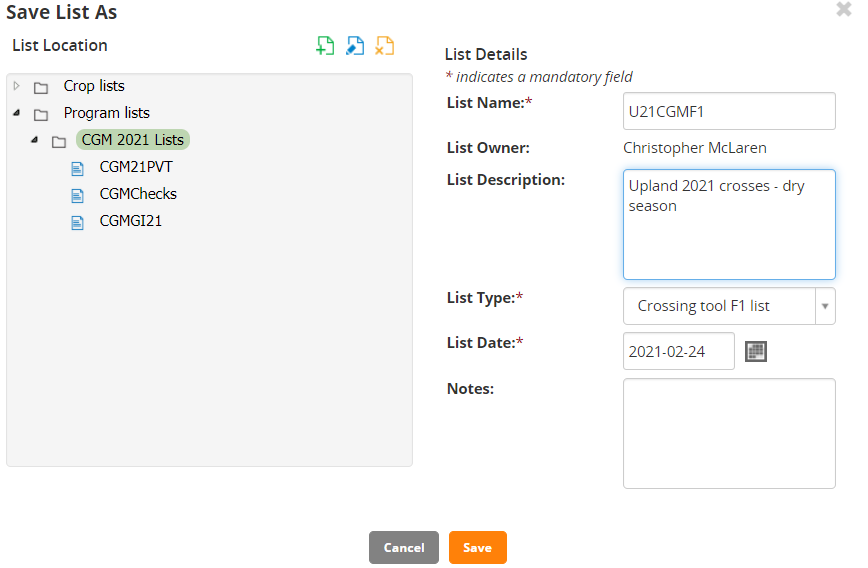
For list details, enter the following
- Enter "U21<your initials>F1" as list name
- Enter "Upland 2021 Crosses Dry Season" for description
Click Save. A message box will be shown about saving the crosses.
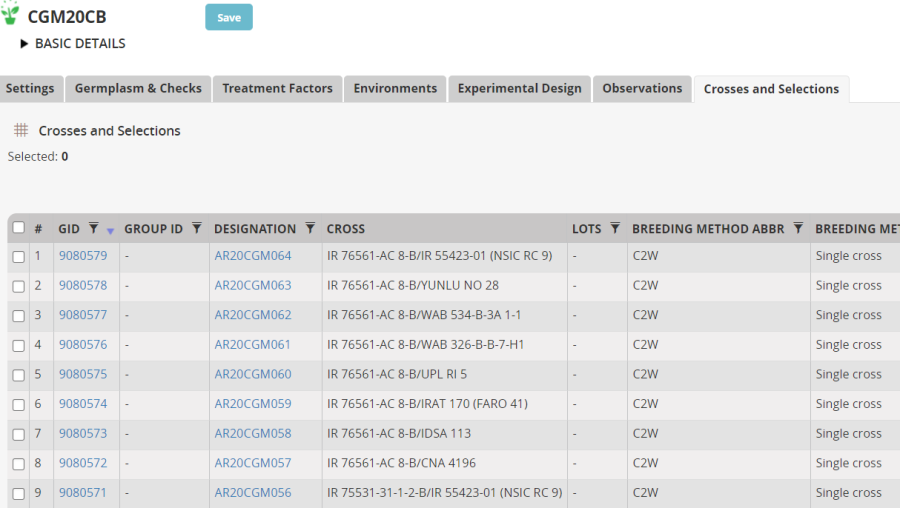
The details about the created list will also be shown in the Crosses and Selections Tab.
Using the Crossing Template to make crosses
The other method of specifying crosses is to use a crossing template. We will show this option in the same nursery as the one where we demonstrated the Crossing Tool although generally you would use one method or the other but not both in the same nursery.
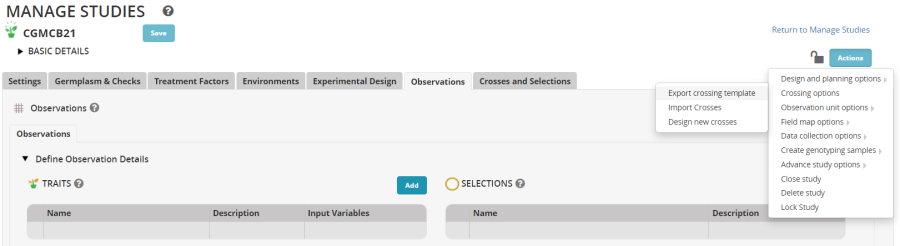
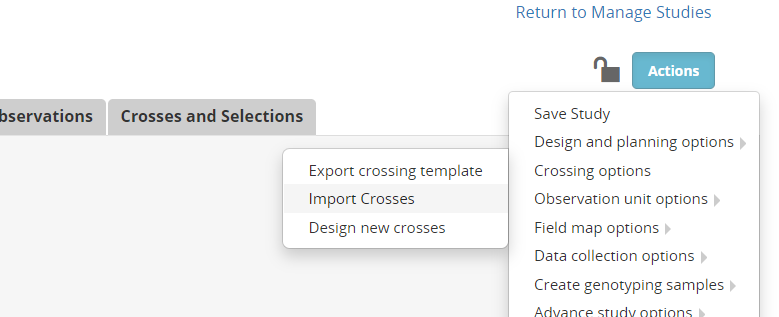
Open the study you created at the beginning of this tutorial (CGM21CB for me). To obtain a crossing template select Actions>Crossing options>Export crossing template:
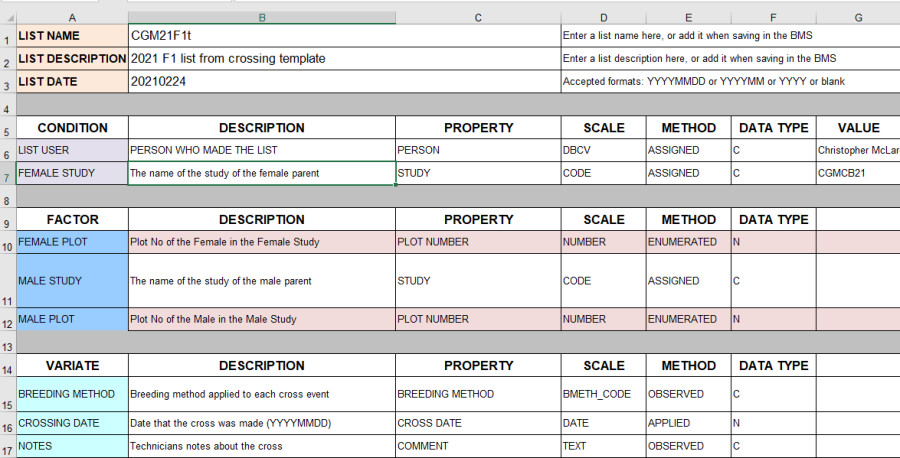
The Crossing Template is an excel file with four sheets. The first sheet is the description sheet and you can fill in some metadata about the F1 list you wish to create, such as a list name <your initials>21F1t, a description "2021 F1 list from crossing template" and a date.
The second sheet is the observation sheet and this is where we will specify the crosses we make.
The third sheet is a Codes sheet where we can look up user names for the description metadata and breeding method codes for the observation sheet. The fourth sheet, Study List is just a fieldbook of the planted material showing you what germplasm was planted on what plot.
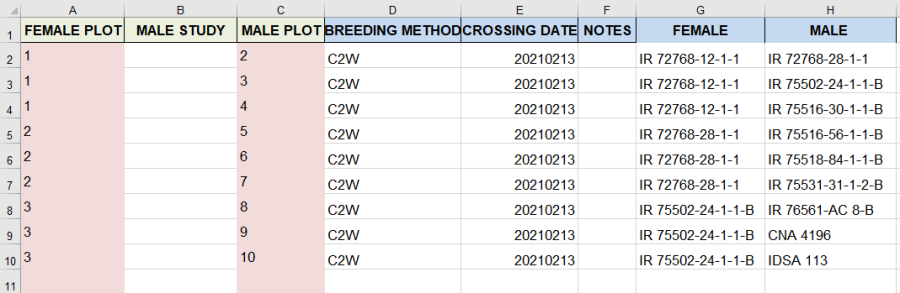
On the observation sheet, the important columns are the FEMALE PLOT and the MALE PLOT. These are the plot numbers in the field layout (shown on the Study list sheet) which are crossed. You can either fill these columns as you do the crosses when the plants are flowering, or before you do the crossing as a specification of the crosses you want to make. The column MALE STUDY allows you to specify another study in the field from where pollen was collected. In this case the MALE PLOT is the plot in that study from where the pollen came. IF MALE STUDY is blank it is assumed to be the same study as the FEMALE PLOT.
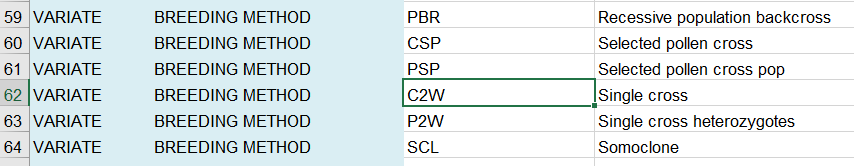
You can fill in the BREEDING METHOD column with the code for the method of crossing you are doing which you look up in the Codes sheet. C2W is the code for a single cross:
You can leave this column blank if you like and it will be filled by BMS.
Finally a CROSSING DATE is required, and NOTES may be useful. The two columns Female and Male are not required, they do not come with the template, but have been added here to show what crosses the plot pairs are producing. The have been filled with the excel functions VLOOKUP. (Optional)
=VLOOKUP(A2,'Study List'!$B$2:$F$17,5,FALSE) for Female and =VLOOKUP(C2,'Study List'!$B$2:$F$17,5,FALSE) for Male. Of course, this will not work for the Male if the pollen came from another study.
Once the crossing template is complete (at the end of the crossing cycle) you can import the crosses by selecting Actions>Crossing options>Import crosses.
Choose the template file. If you have the Female and Male columns you get a warning that these will be ignored. And then you are asked to specify how the breeding method will be supplied.
You can use the method specified in the template (if you specified it as we have), or you can get BMS to chose the method based on parental status, or you can pick the method for a list.
We continue with the default selection.
Now you need to specify a naming convention. We will check the Specify name format option. We will use the same format as we used in the last section: prefix AR20<your initials> with three digits for the sequence code:
Notice that the next name in the sequence will be AR20CGM065 because we have already made 64 crosses with that naming pattern.
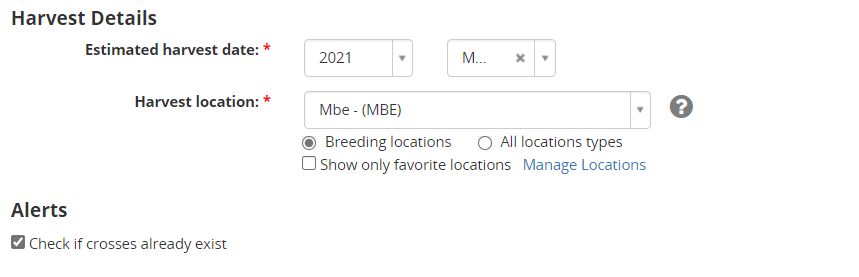
Specify the harvest month and the harvest location and check the box to be warned if the cross already exists (according to parental combination) in the database. Click continue.
You will get a list of crosses to review:
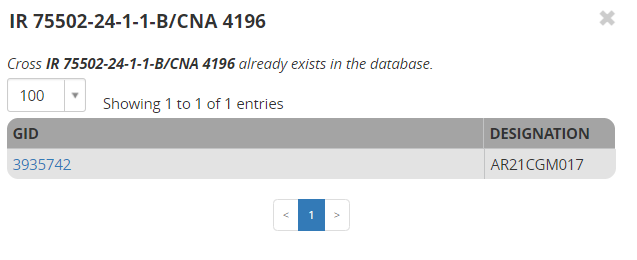
You see in the ALERTS column that we are being warned that two of the crosses have been made before. If you click on View Existing Crosses you will see that those duplicates are crosses I made before (in the previous section):
The option to remove those duplicate crosses would be to select the Omit alerted crosses box. Click Finish and save the list.
You can also see the results in the Crosses and Selections tab where the 9 new crosses have been added to the previous crosses made.
Table 1: BMS Breeding Methods for Self Fertilizing Crops
METHN | MTYPE | MGRP | MCODE | MNAME | MDESC | ||
Methods for storing historical pedigrees with incomplete information | |||||||
1 | GEN | S | UGM | UNKNOWN GENERATIVE METHOD SF | Unknown generative method for storing historic pedigrees for self fertilizing species. | ||
4 | GEN | S | BDU | F1 BACKCROSS, CYTOPLASM UNKNOWN SF | Cross of F1to recurrent parent when the direction of the cross is unknown for storing historic pedigrees for self fertilizing species. | ||
6 | GEN | S | BRU | F2 BACKCROSS, CYTOPLASM UNKNOWN SF | Cross of F2 to recurrent parent when the direction of the cross is unknown for storing historic pedigrees for self fertilizing species. | ||
8 | GEN | G | CCU | CROSS, CYTOPLASM UNKNOWN | Cross between two plants, unknown which is female | ||
31 | DER | S | UDM | UNKNOWN DERIVATIVE METHOD SF | Unknown derivative method in self-fertilizing species: for storing historic pedigrees | ||
Generic Maintenance Methods | |||||||
60 | MAN | G | IDN | PLANT IDENTIFICATION | Identifying and naming a plant or population. | ||
61 | MAN | G | NSI | SEED INCREASE | Increase seed of a cultivar, line, population or accession. | ||
62 | MAN | G | ISE | IMPORT | Import seed, clones or tissue culture of a cultivar, line, population or accession. | ||
63 | MAN | G | ESE | EXPORT | Export seed, clones or tissue culture of a cultivar, line, population or accession. This method is not required. | ||
64 | MAN | G | SSN | STORE SEED NORMAL | Store seed of a cultivar, line, population or accession in normal method: drift not expected. It is unlikely that this method is needed. | ||
65 | MAN | G | SSM | STORE SEED MEDIUM TERM | Store seed of a cultivar, line, population or accession in medium term storage. Some genetic drift is expected. Storage is between 0-4OC and low RH. | ||
66 | MAN | G | SSL | STORE SEED LONG TERM | Store seed of a cultivar, line, population or accession. Genetic drift is expected. Storage is about -18OC. | ||
Generative Methods for Inbreeding Crops | |||||||
101 | GEN | S | C2W | SINGLE CROSS | Cross between two single plants. If both parents are fixed (pure) inbred lines there will be no segregation for gametes or genotypes and theoretically all crosses will result in the same genetic outcome. In plant breeding practice the theoretical situation is rarely encountered. In spite of this the usual practice is to bulk the seed. However, in genetical studies it is often necessary to keep individual seed separate. When this is done a separate entry in the germplasm table is required for each entity (seed) kept separate. | ||
102 | GEN | S | C3W | THREE-WAY CROSS | Cross between two plants, one an inbred line and one a single cross (usually an F1) and thus segregating for gametes. In the theoretical case, rarely achieved, the inbred line would be fixed and the F1 a cross between fixed lines. The segregation for gametes results in different genetic outcomes among different progeny, hence a number of crosses using the same F1 is usually made. Since different F1 s are genetically the same (theoretically) only one F1 is required. In plant breeding programs the different crosses are usually bulked. Again, if individual seeds are kept separate a different entry is required in the germplasm table. | ||
103 | GEN | S | CDB | DOUBLE CROSS | Cross between two single crosses (usually two F1s) and hence both segregating for gametes. The comments for method 102 apply but now for both female and male sides of the cross. Again, if individual seeds are kept separate a different entry is required in the germplasm table. | ||
104 | GEN | S | CFT | FEMALE COMPLEX TOP CROSS | Cross between a female inbred line and a three-way or more complex cross among inbred lines, thus the male is segregating for genotypes as well as gametes. A consequence of the genotypic segregation is that selection can, and is usually made among the plants used as male parents. A consequence is that there will be genetic variation both within and between each cross. Usually all seed is bulked and selection practiced among the progeny. A different entry is required in the germplasm table for each entity kept separate. | ||
105 | GEN | S | CMT | MALE COMPLEX TOP CROSS | Cross between a male inbred line and a three-way or more complex cross among inbred lines, thus the female is segregating for genotypes as well as gametes. The same genetic consequences occur as for the previous complex cross except for the cytoplasm. This method is rarely if ever encountered in practice because of the difficulty of using many females. A different entry is required in the germplasm table for each entity kept separate. | ||
106 | GEN | S | CCX | COMPLEX CROSS | Cross between two three-way or more complex crosses among pure lines, thus both sides are segregating for both gametes and genotypes. A different entry is required in the germplasm table for each entity kept separate. | ||
107 | GEN | S | BC | BACKCROSS | Backcross to recover a specific gene. The coding in the genealogical table records which parent was used as the female in each cycle. A different entry is required in the germplasm table for each entity kept separate. | ||
108 | GEN | S | BCR | BACKCROSS RECESSIVE | Backcross to recover a recessive gene. As this requires a self fertilization (derivative method) in the process some ICIS administrators may distinguish this as a separate method. A different entry is required in the germplasm table for each entity kept separate. | ||
109 | GEN | S | CIS | INTERSPECIFIC CROSS | Cross between two species. The problem with making this a separate method is that the species cross could be made by any of the previous (101-108) or following (110-113) methods. | ||
110 | GEN | S | CSP | SELECTED POLLEN CROSS SF | A bulk of pollen from a selected set of males used to pollinate a female inbred line. | ||
111 | GEN | S | CRP | RANDOM POLLEN CROSS SF | A random bulk of pollen from some population used to pollinate a female pure line. Male is then a population and will be recorded as a single entity. | ||
112 | GEN | S | CGO | OPEN POLLENATED SF | Open pollination in a self- fertilized species | ||
151 | GEN | S | MUN | NATURAL VARIANT SF | A recognized naturally occurring variant in a self- fertilizing population. | ||
152 | GEN | S | MIP | INDUCED MUTATION POPULATION SF | A population derived from inducing mutation in a inbred line. | ||
153 | GEN | S | SCL | SOMACLONE SF | Variation induced through tissue culture of a inbred line. | ||
154 | GEN | S | ALP | ALLOPOLYPLOID SF | Polyploid formed by doubling the chromosomes of a cross between two or more species. Wheat is an allopolyploid as it contains genomes from three different species. | ||
155 | GEN | S | AUP | AUTOPOLYPLOID SF | Polyploid formed by doubling the chromosome number of a species. Lucerne (alfalfa) is an autopolyploid with 4 sets of the same genome. | ||
156 | GEN | S | HAP | HAPLOID SF | Individual with chromosome content of reduced gamete. Often formed by female progenitors crossed with a haploid inducer. | ||
157 | GEN | S | TRN | TRANSGENIC NUCLEUS SF | Individual derived from genetic transformation of the nucleus in a self fertilizing species. | ||
158 | GEN | S | TRC | TRANSGENIC CYTOPLASM SF | Individual derived from genetic transformation of a cytoplasm inclusion (e.g. chloroplast) in a self- fertilizing species. | ||
Derivative Methods for Inbreeding Crops | |||||||
201 | DER | S | MIL | INDUCED MUTATION LINE | A recognized mutation selected from an induced mutation in a line of a self-fertilized species. | ||
202 | DER | S | DDH | DOUBLE HAPLOID LINE | Individual produced by doubling haploid individual usually by anther culture in a self- fertilized crop. | ||
203 | DER | S | DPR | PURIFICATION | Selection of one or a few plants from an inbred line or pure line cultivar. | ||
204 | DER | S | DRU | ROUGING SF | Eliminating off types from a inbred line or pure line cultivar. | ||
205 | DER | S | DSP | SINGLE PLANT SELECTION SF | Derivation through selection of a single plant, inflorescence, fruit or seed from a self-fertilizing population. | ||
206 | DER | S | DSB | SELECTED BULK SF | Derivation through bulking seed from a selected set of single plants from a self-fertilizing population. | ||
207 | DER | S | DRB | RANDOM BULK SF | Derivation through bulking seed from a random selection of single plants from a self-fertilizing population. | ||
208 | DER | S | DSD | SINGLE SEED DESCENT SF | Derived through the production of a single individual without selection from each individual in a segregating population. | ||
209 | DER | S | DRS | CMS RESTORER SELECTION | Restorer Lines selected at the end of a program to back cross a gene which restores male fertility to lines carrying a Male Sterile Cytoplasm (CMS) to the male of a commercial hybrid. | ||
210 | DER | S | DMS | CMS MAINTAIN ER SELECTION | Maintainer line selected at the end of a program to create the male fertile equivalent of the CMS female parent of a hybrid | ||
251 | DER | S | ALP | LANDRACE POPULATION SF | Acquisition only. | ||
252 | DER | S | ALL | LANDRACE LINE SF | Acquisition only. | ||
253 | DER | S | ALC | LANDRACE CULTIVAR SF | Acquisition only. | ||
254 | DER | S | ACP | COLLECTION POPULATION SF | Acquisition only. | ||
255 | DER | S | ACL | COLLECTION LINE SF | Acquisition only. | ||
256 | DER | S | AWP | COLLECTION WILD SPP POPULATION SF | Acquisition only. | ||
257 | DER | S | AWL | COLLECTION WILD SPP LINE SF | Acquisition only. | ||
258 | DER | S | ADP | COLLECTION WEEDY SPP POPULATION SF | Acquisition only. | ||
259 | DER | S | ADL | COLLECTION WEEDY SPP LINE SF | Acquisition only. | ||
Management Methods for Inbreeding Crops | |||||||
301 | MAN | S | NSP | SEED INCREASE PLANT SF | Seed increase from a single plant in a self-fertilized species. | ||
302 | MAN | S | NMX | SEED INCREASE MIXTURE SF | Seed increase from a number of selected plants in a self- fertilized species. | ||
303 | MAN | S | NBK | SEED INCREASE BULK SF | Seed increase from an unselected bulk in a self-fertilizing species. | ||
320 | MAN | S | VPL | PURE LINE FORMATION | Forming a pure line CV in a self-fertilizing species. | ||
321 | MAN | S | VHY | HYBRID FORMATION SF | Forming a hybrid CV in a self-fertilizing crop. | ||
322 | MAN | S | VML | MULTI-LINE FORMATION SF | Forming a multi-line CV in a self-fertilizing crop | ||
323 | MAN | S | VBS | BREEDERS SEED SF | Producing Breeder's Seed. Pure seed produced by breeder (usually some kept by breeder) in a self-fertilizing crop. | ||
324 | MAN | S | VFS | FOUNDATION SEED SF | Producing Foundation Seed. Pure seed derived from Breeders seed (usually kept by seed producing organization) in a self-fertilizing crop. | ||
325 | MAN | S | VCS | CERTIFIED SEED | Producing Certified Seed. Pure seed produced under supervision by Government Protocols. | ||
326 | MAN | S | VCR | CULTIVAR RELEASE | Release a cultivar |