Once the study design has been generated or imported, the Observations table is populated with independent study variables and sub-observation datasets can be created to record repeated measures.
The rows of the Observations table represent the experimental units, which are randomized or non-randomized depending on design. Experimental units can be defined in many ways: plots, pots, individual plants, fruits, ect. The manual will use "plots" to generically describe the highest level of observation, as this is the most common experimental unit in breeding.
Saving the trial will paginate the Observations table by environment. Select any environment to review the plot details.
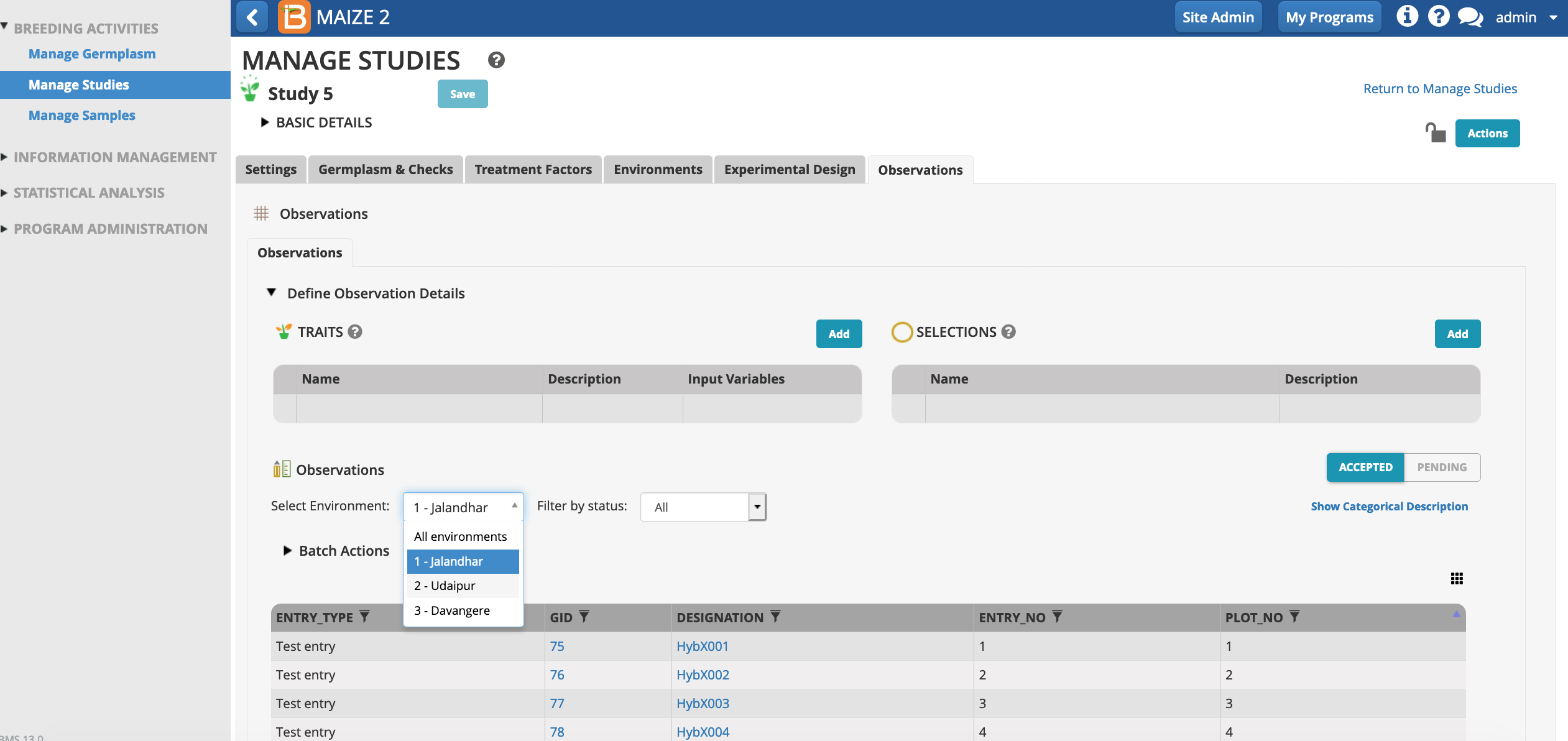
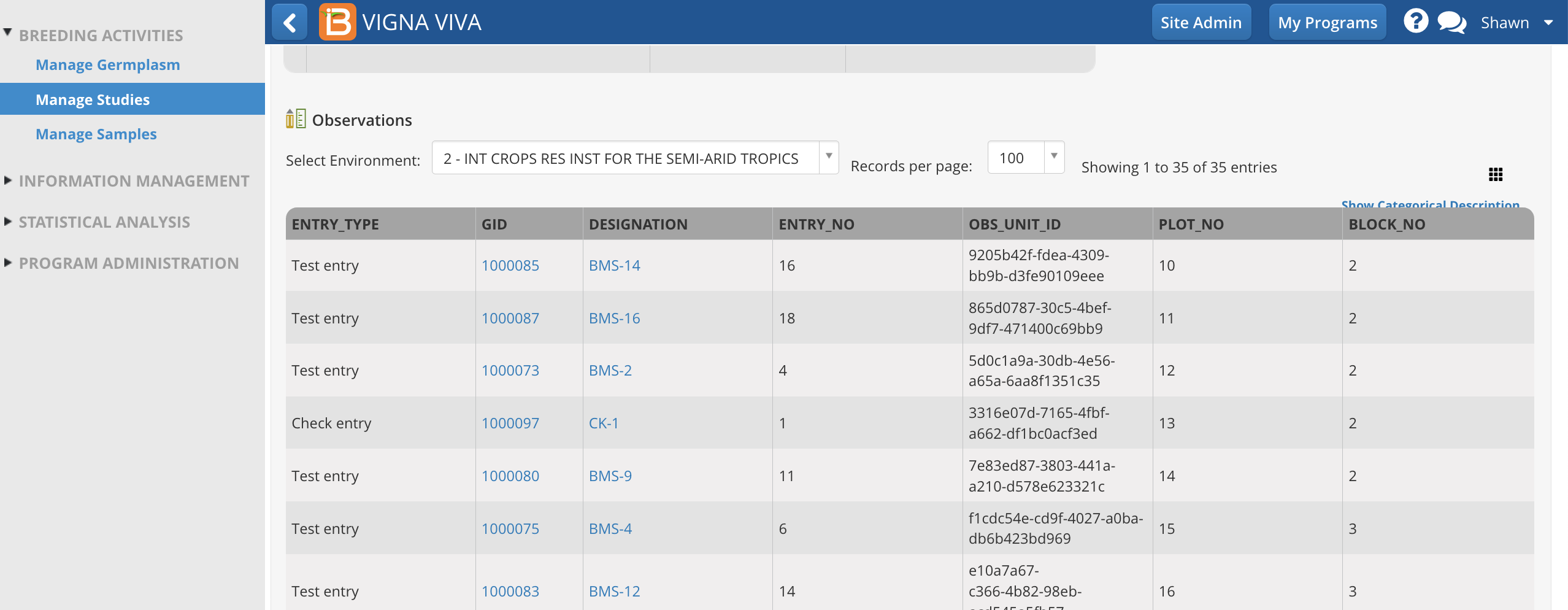
Independent variables can be hidden and revealed in the tabular user interface. One important column, OBS_UNIT_ID, is hidden by default, because it is not meant to be human readable. OBS_UNIT_ID is an alphanumeric sequence designed for data capture that uniquely identifies the observation. The OBS_UNIT_ID is appropriate for barcoding the observation unit (plot, plant, pot, ect...) when the Study Book file is exported
Make columns of data by visible/hidden by selecting the dotted rectangle.
OBS_UNIT_ID is revealed in the user interface after the selection.
Traits and trait aliases are defined by the crop ontology. If you do not find a trait of interest from the drop down menu, see Manage Ontology for instructions on adding new traits.
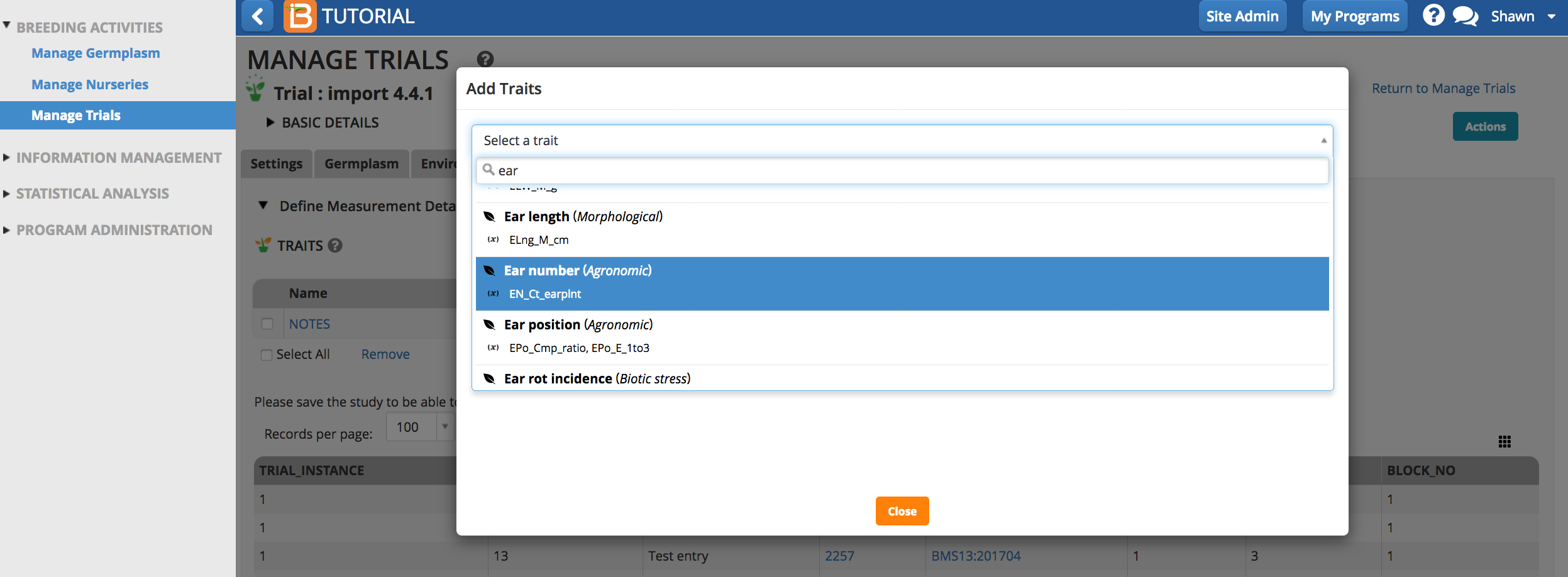
Select the Add button to specify traits to measure (the dependent variables). Type a word or part of a word that describes the trait that will be measured. You can search by name or the alias of the variable.
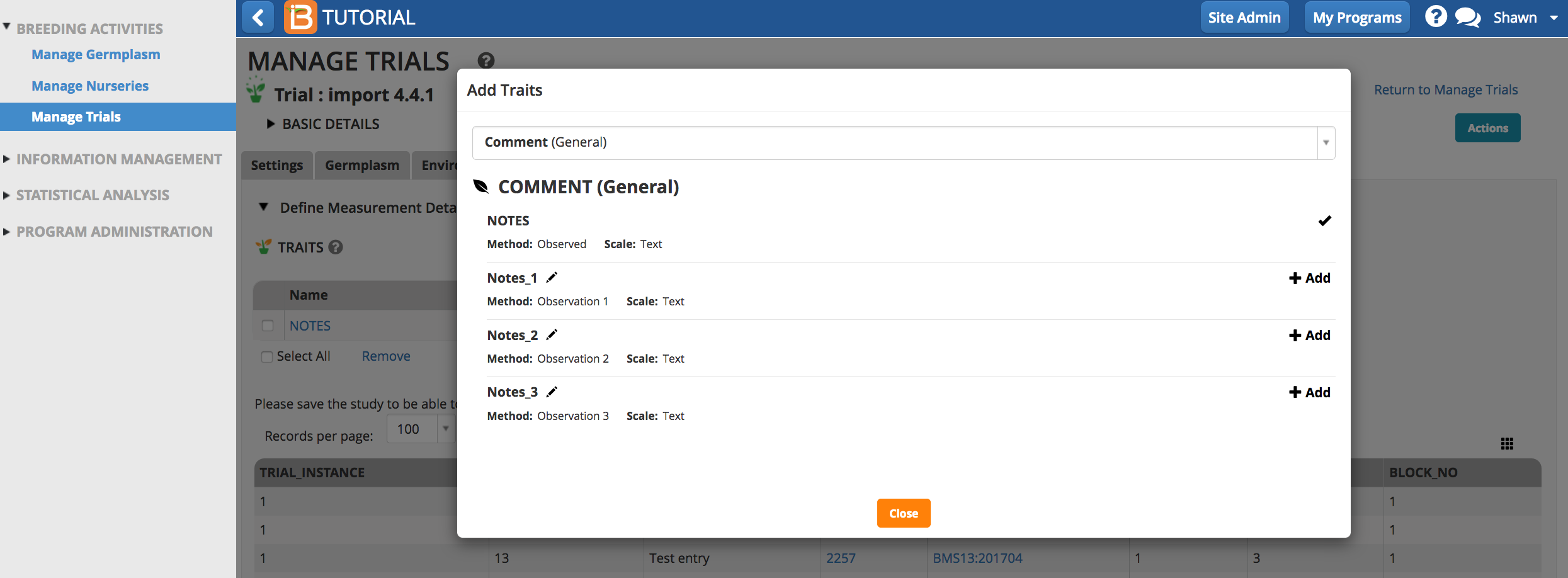
Once selected, the traits of interest will appear as an empty column of data in the measurements table.
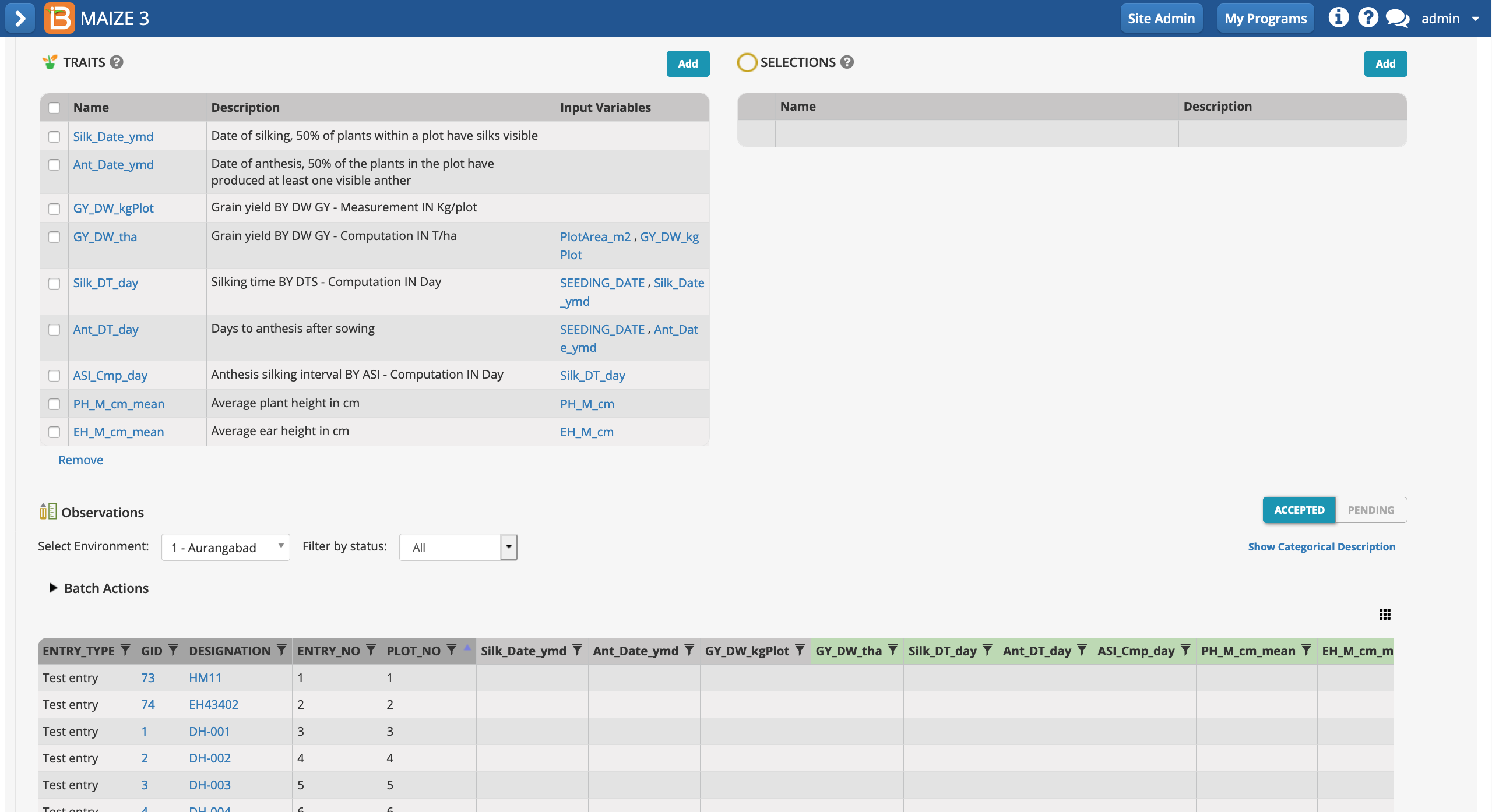
The saved study is ready for (1) data collection and/or the creation of a (2) sub-observation dataset to record repeated measures.
If you want to advance within plot selections, a selection variate can be added to the study (see more on Selections).
Select Add Selections. Choose selection variate. Close popup.
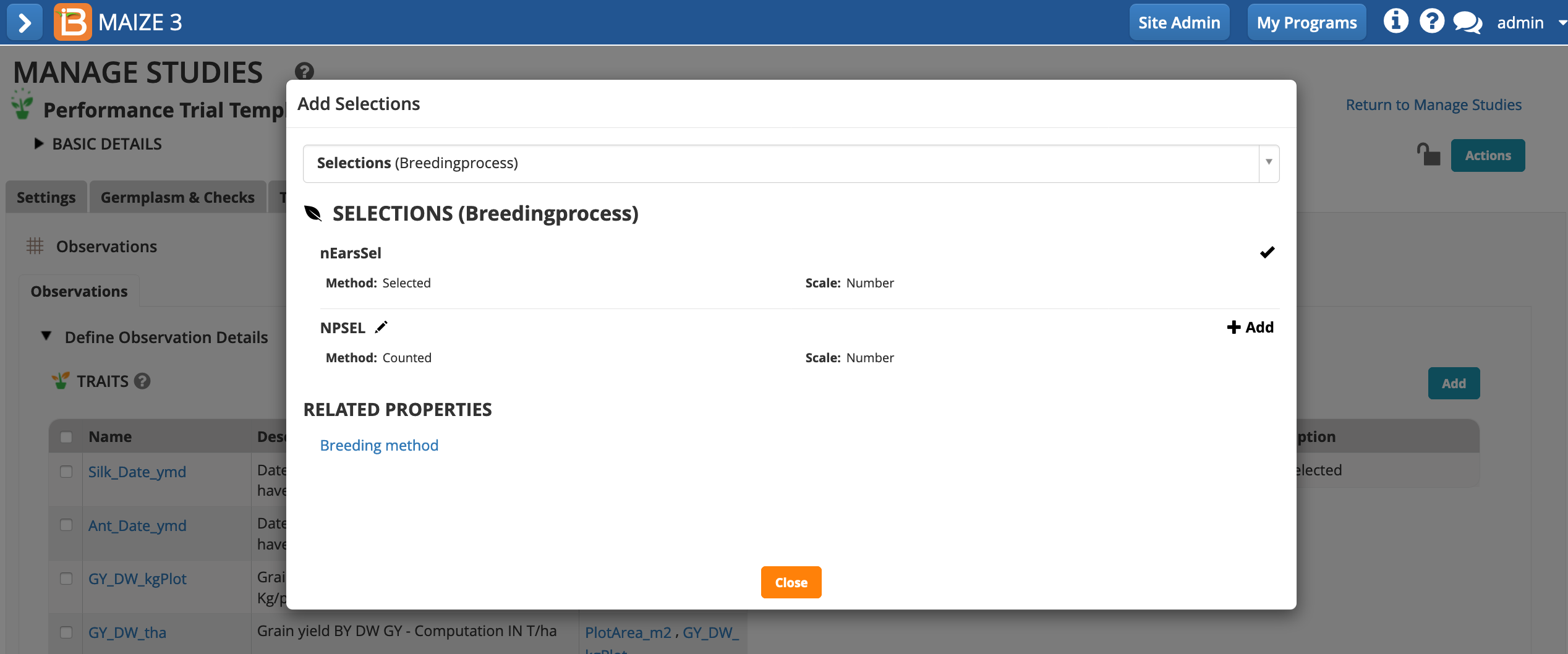
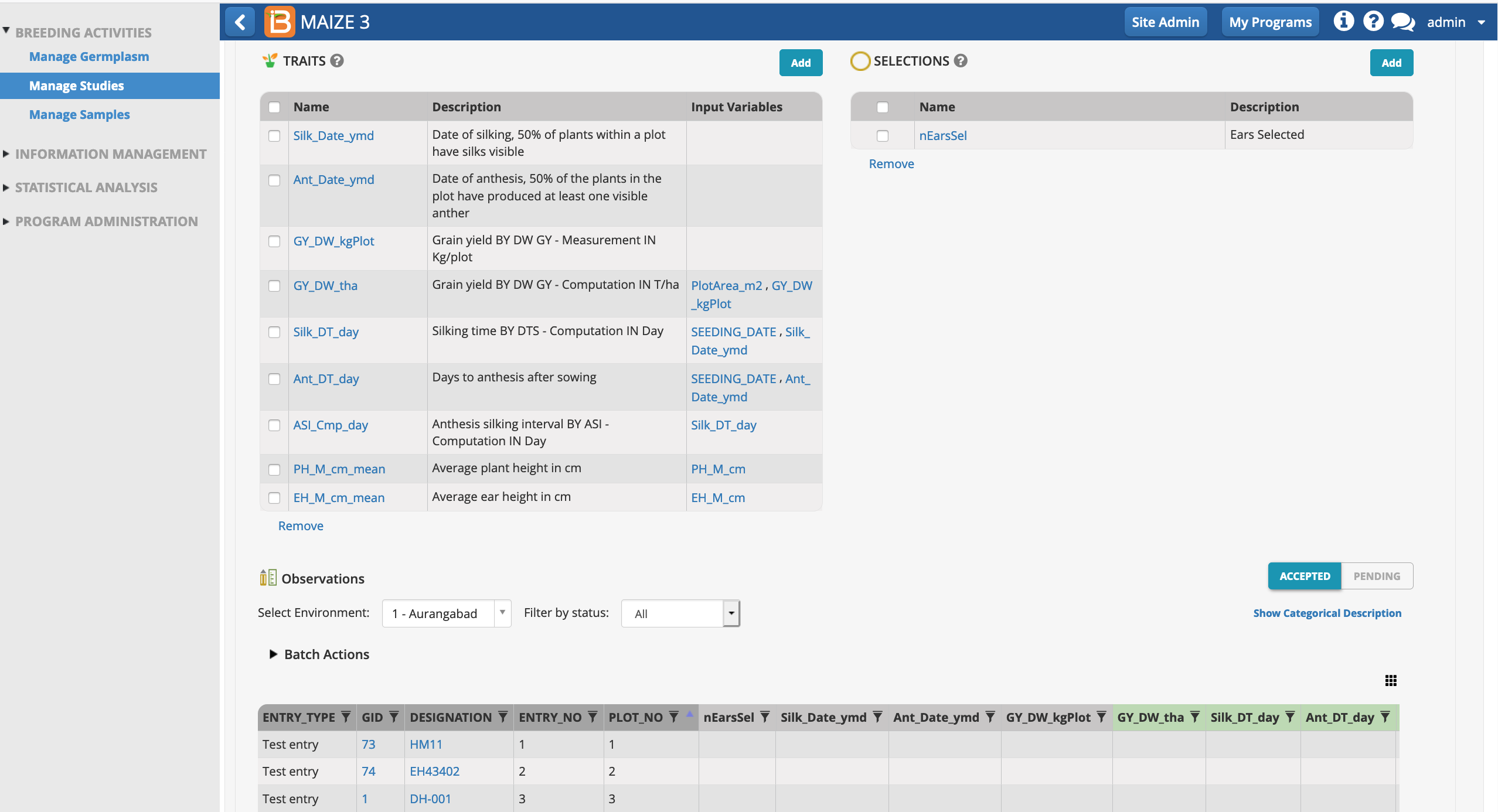
The selection variate is included as a column in the measurements table, and is ready to be filled with the number of ears selected from each plot.
Files column is available in Observations and Sub observations tabs.
Note: The file extensions supported: png, jpeg, jpg, pdf, json, csv, mpeg, tiff, bmp, and gif.
Select a plot
Hovering a cell in the File column shows the file icon.

Click on it and the File Manager will be displayed
Browse a file, a preview is generated in this step.
Optionally you can associate a variable to the file > Select the variable (only traits and selection variables are displayed in the drop-down)

Upload. A success message is displayed, the file is added to the Files table, and the associated variable is added in the variables column.
Clicking in the file row you could see the preview again.
Click on the Download icon
Click on the trash icon for deleting a file. A confirmation message will be displayed

Confirm. The file is not displayed anymore.
Once the files are loaded in the Observations/Sub-observations you can open the File Manager from the Files column or from a variate column:

Once experimental design has been generated and the Observation table established, you are able to create additional data collection tables for repeated measures (sub-observations units).
Common repeated measures include:
Measure individual plants within a plot
Measure different quadrats within a plot
Measure the same trait at multiple time point
In the following example, a maize breeder is planning to measure the height of 5 plants per plot at maturity.
Select Plants as the sub-observation units.
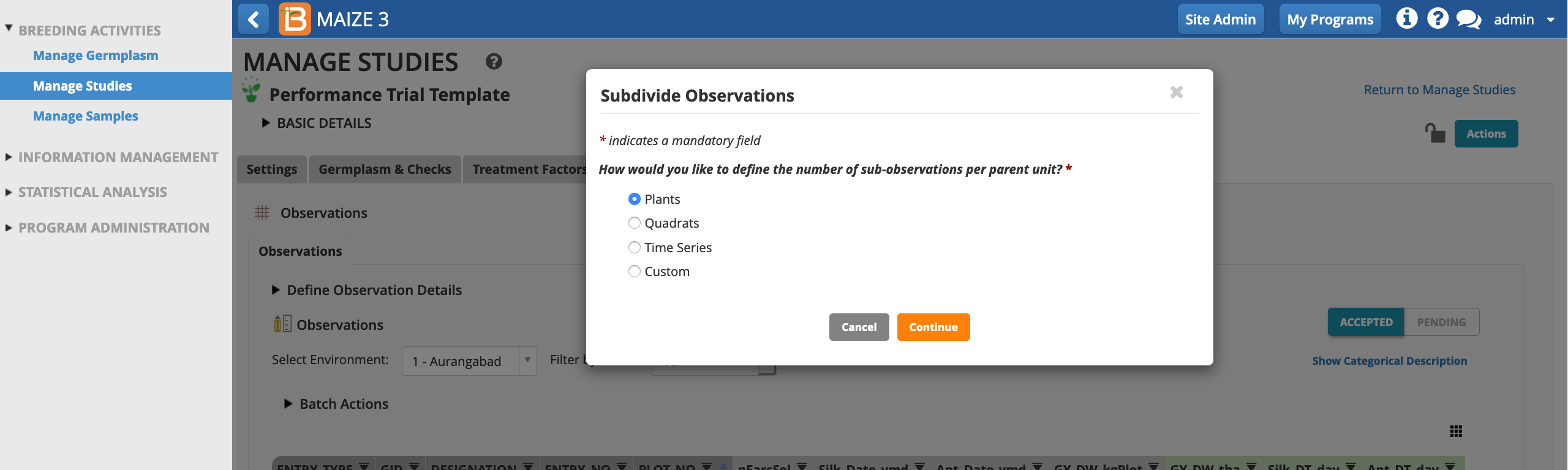
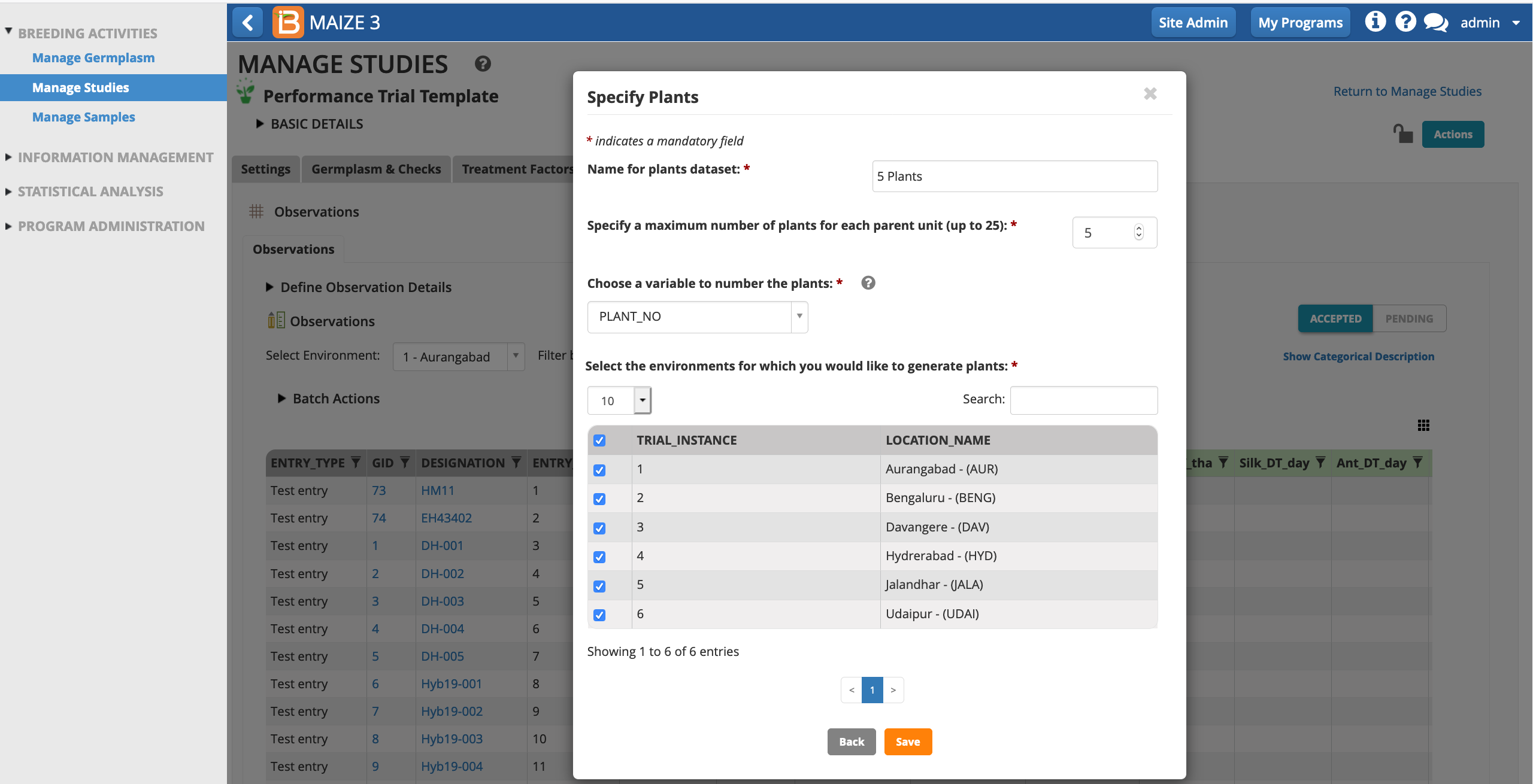
Give the sub-observation data set a unique name. Specify 5 plants per plot. Leave PLANT_NO as the default numbering variable. Choose to sub-sample all study locations and Save.
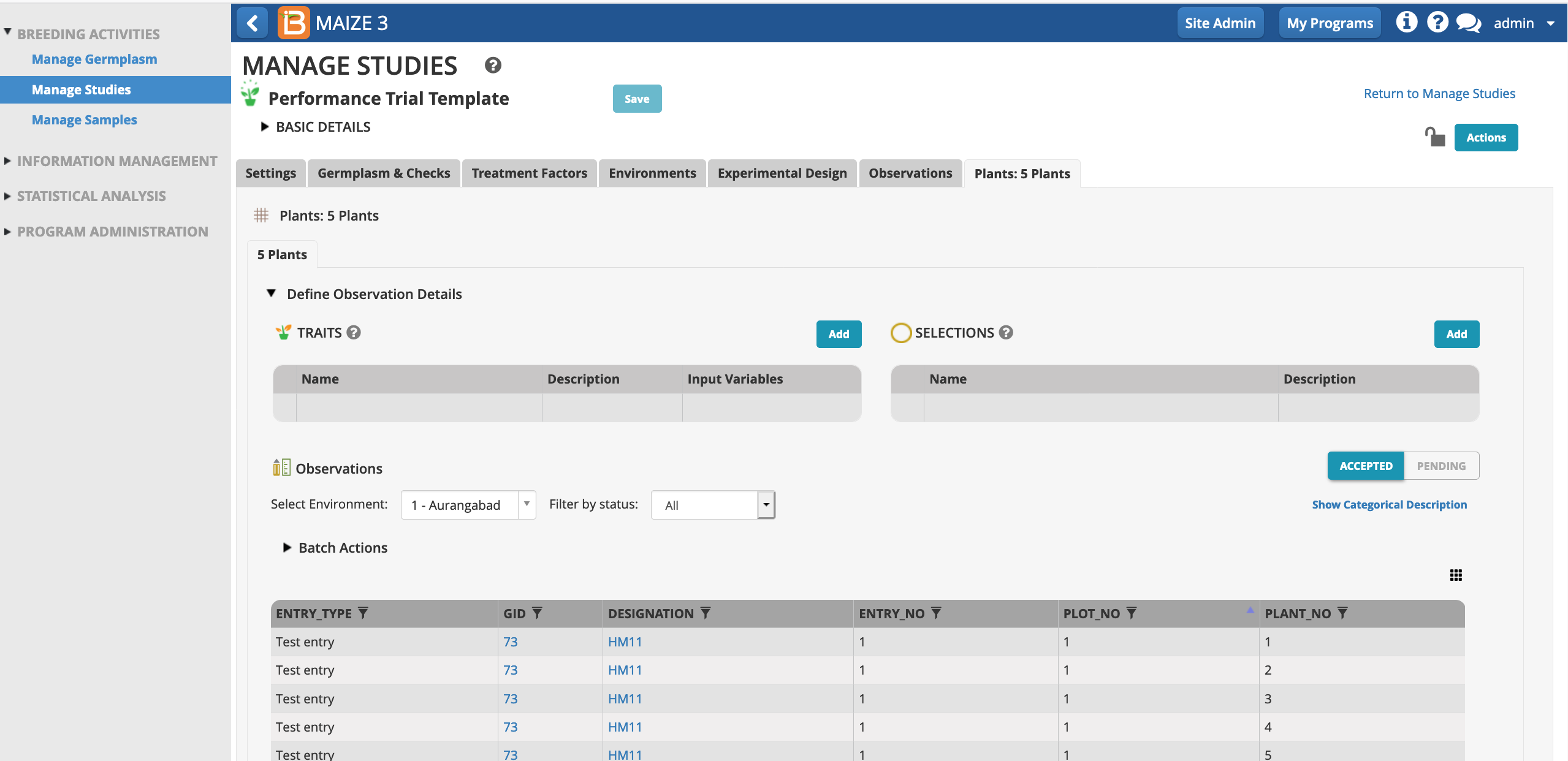
Select Add to add plant level trait details.
Each plot now contains 5 rows corresponding to 5 plants per plot.
Select the traits and/or selection variables of interest to include as columns in the sub-observation dataset.
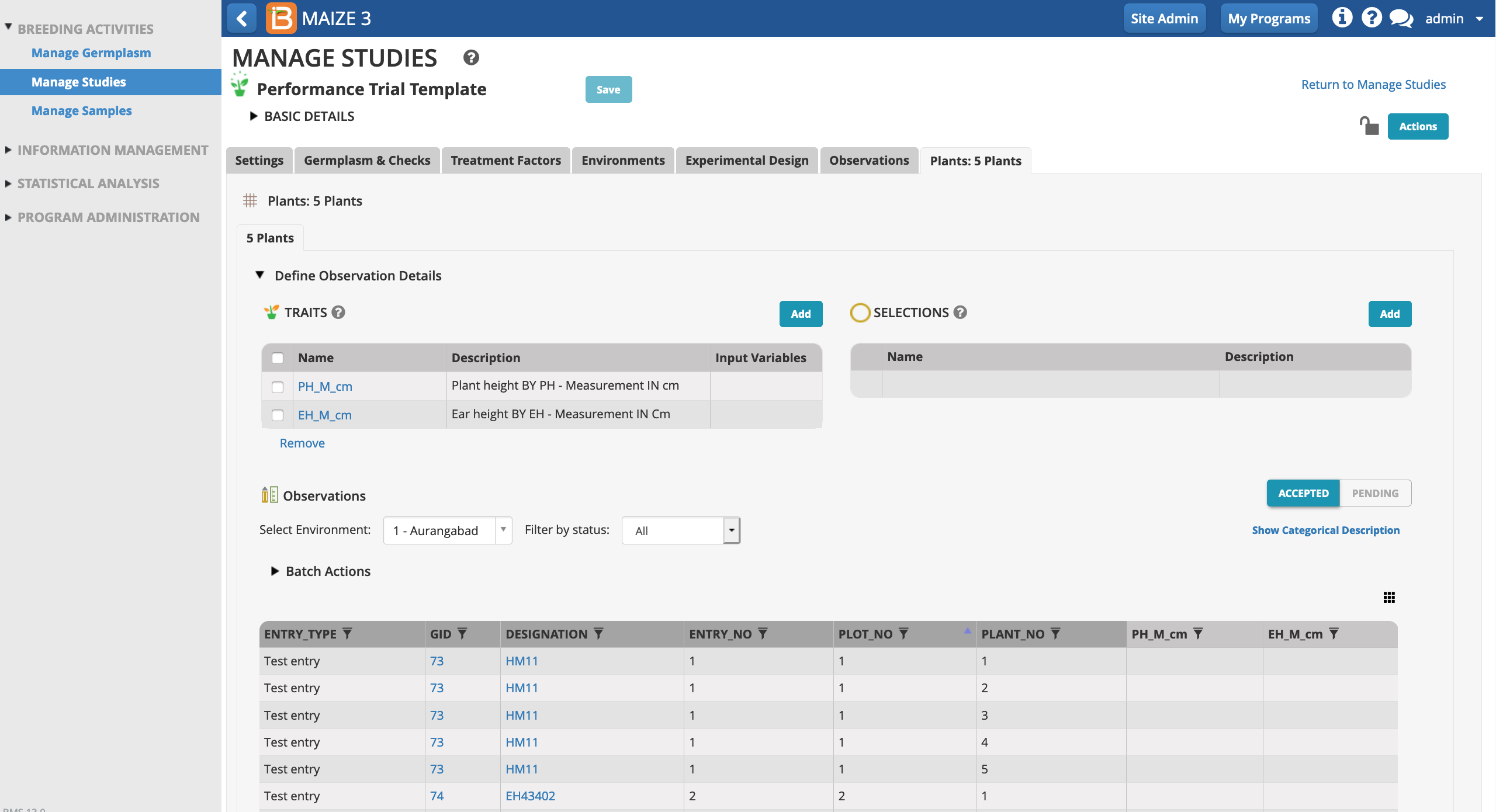
In this sub-observation dataset plant and ear heights (cm) will be measured from 5 representative plants in each plot. Each plant in the sub-observation dataset receives a unique OBS_UNIT_ID (see above for more info) suitable for barcoding individual plants within the plot.
In the following example, a maize breeder is planning to gather ears of interest from experimental plots to take ear-specific measurements. The breeder doesn't know in advance how may ears will be collected, but expects to collect no more than 7 per plot.
'Ears' is not a default sub-sampling option. Select the Custom option and Continue.
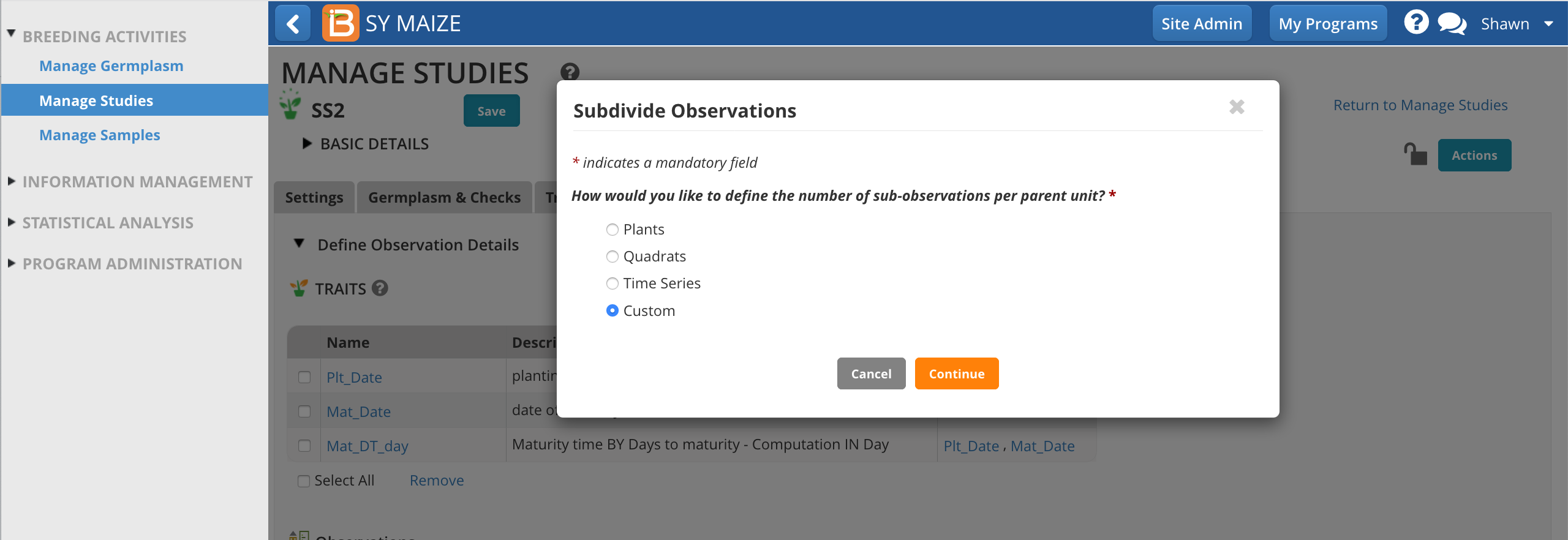
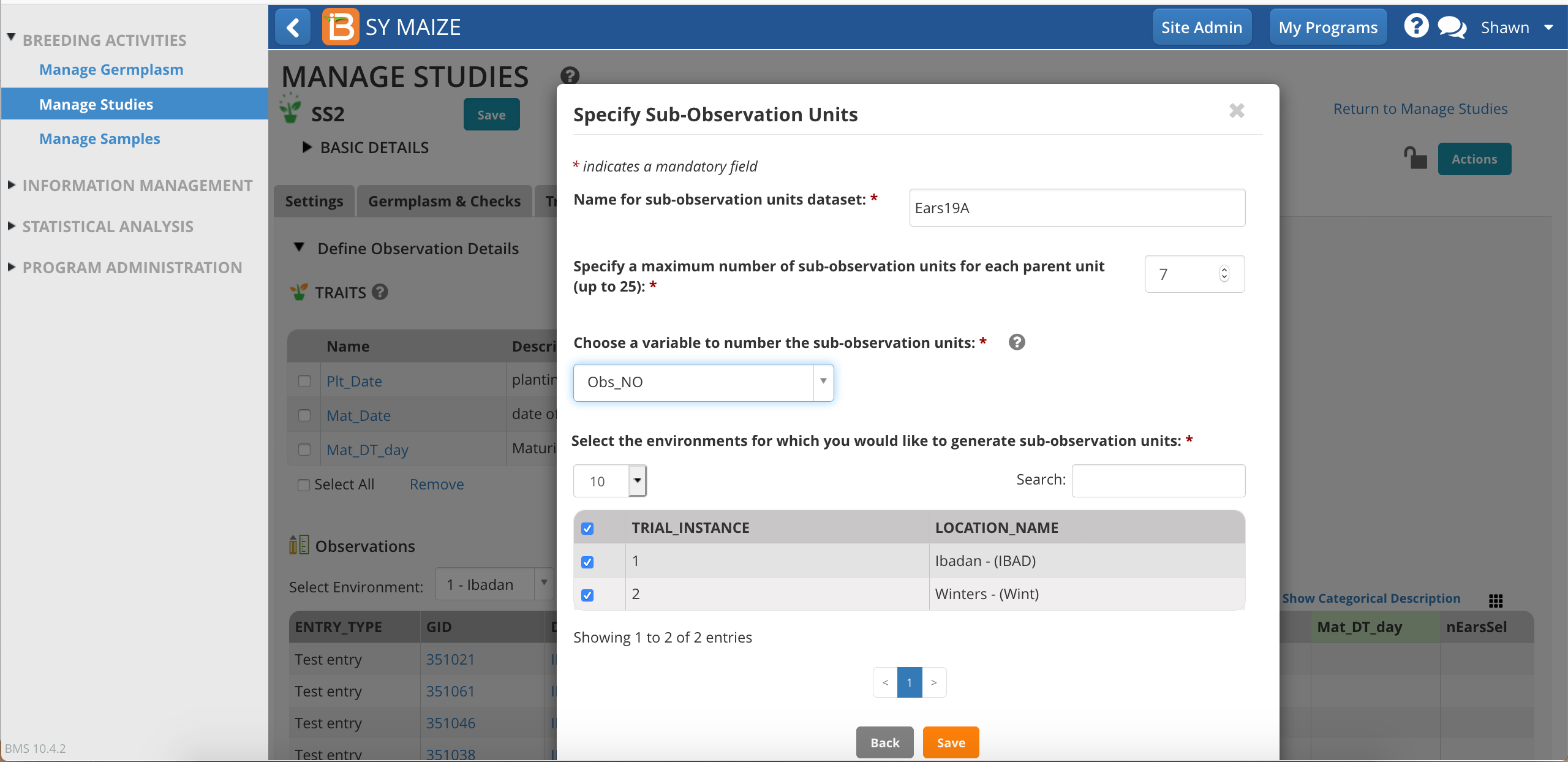
Give the sub-observation data set a unique name.
Specify 7 ears as the maximum collected per plot.
Choose a numbering variable. In this case, Obs_NO. Note: custom sub-observations units will not have a corresponding numbering variable until you create one (see details below).
Choose to sub-sample both study locations.
Save.
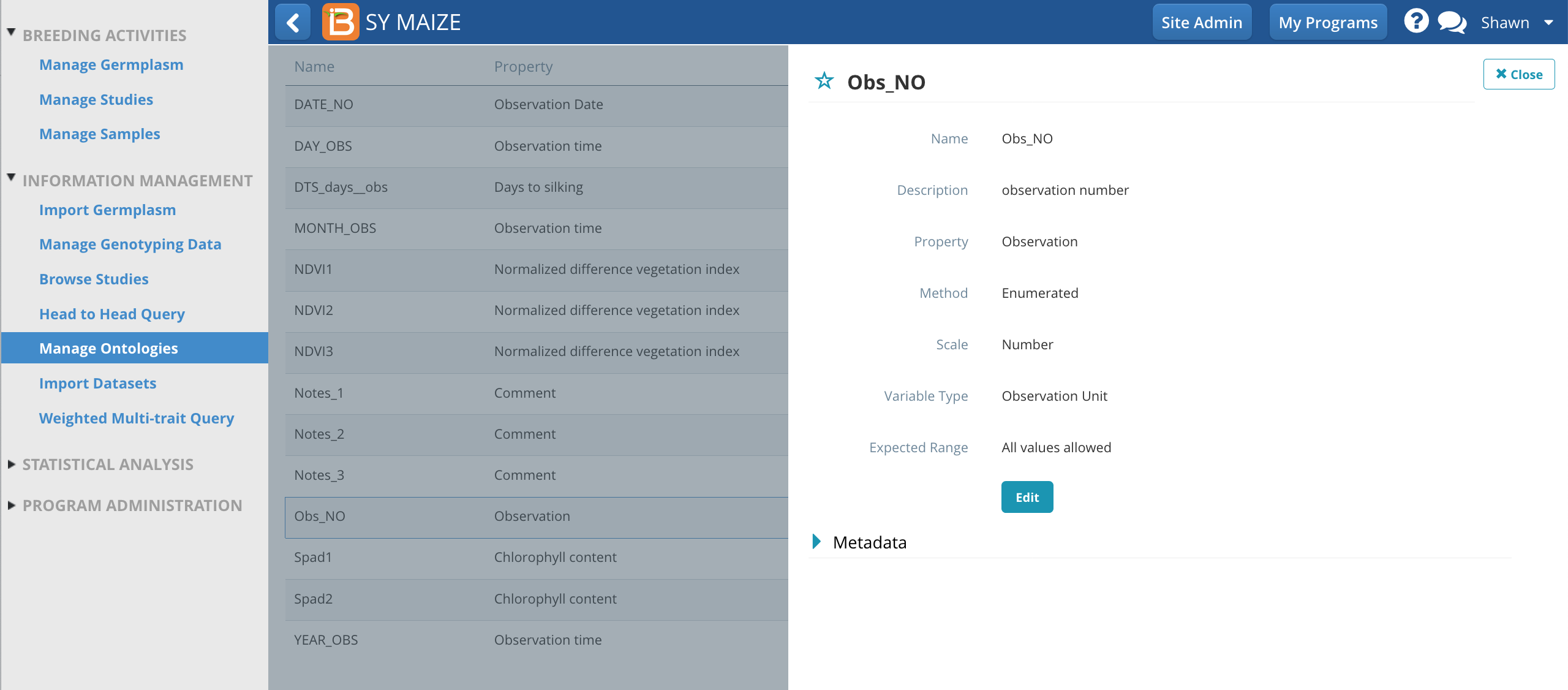
Customize Observation Unit Variable
The observation unit variable, Obs_NO, provides a generic way to number any observation. Alternatively a more specific term, like EAR_NO, could be created via Manage Ontologies.
The Ears sub-observation dataset is now ready to accept 7 ear measurements per plot. Add ear traits to the dataset and Save.

There are cases where the breeders realize at the planting stage that there is not enough seed for all the plots they have in the experimental design. A tradeoff must be done in such cases and the breeder might opt for 1) Replace the entry for all plots 2) Replace the entry for a particular plot 3) Add a new entry as a replacement for a particular set of plots.
With the Change plot entry option, you can 1) Replace the entry for all plots and 2) Replace the entry for a particular plot
Select one or more than one plot in the observations table
Notice in the screen displayed: 1) All environments are selected and, 2) a filter by GID is used. These selections permit the replacement of the entry for all plots in the study.

Actions > Observations unit options > Change plot entry
The Change plot entry screen is displayed. The number of plots selected for the number of environments is provided.
All germplasm used in the study is listed. The columns and information of the table correspond to the germplasm and checks view.
Select an entry to perform the replacement

Confirm
A confirmation message is displayed

OK
The system replaces the selected entry in all of the selected plots updating all the germplasm-specific information (gid, designation, etc) and entry information (entry type, entry code, etc).
These changes propagate to the sub observations if present.
NOTE: You could not proceed with the replacement if i) the study has an associated MEANS dataset, or ii) new germplasm has been generated (through advances or crosses) within the study, or iii) or one or more of the selected plots has genotyping samples associated to it, or iv) one or more of the selected plots has pending or confirmed inventory transactions associated to it. A proper error message will be displayed.