...
Often an existing trial may be available in excel such as the file CGM20AVT.xlsx where the experimental design and the data are stored in a format like the one shown below.
There may be data for several sites stacked together as with the file CGM20AVT.xls (4 sites) which you can find in the Sample Files folder, or the data for different sites may come is separate but similar files. We need to load this data into a BMS study for analysis.
The first thing to do is to create a germplasm list for the distinct entries in the trial. This can be done by finding the entries in the BMS and adding them to a list with the List Manager, or it can be done by extracting the distinct entries from the file(s) and importing them with Germplasm Import.
We will use Germplasm Import, but it is important to check that the entry numbers and the designations are consistent across every rep of the trial. When you have checked this you can extract the ENTRY_NO and DESIGNATION for site 1 rep 1 and paste it into a Germplasm Import template:
The observation sheet looks like this:
And the description sheet like this:
This list is now imported int BMS taking care to select existing enteries entries wherever appropriate: Go to Import Germplasm, browse for the template file, fill in the import details and click Finish.
Select appropriate germplasm when there are multiple hits, such as for the checks IR 64 and IR 72. (Select the ones with seed stock):
Save the list:
| Anchor | ||||
|---|---|---|---|---|
|
...
Now open the Experimental Design tab and click import an experimental design:
Browse to the layout file and open it:
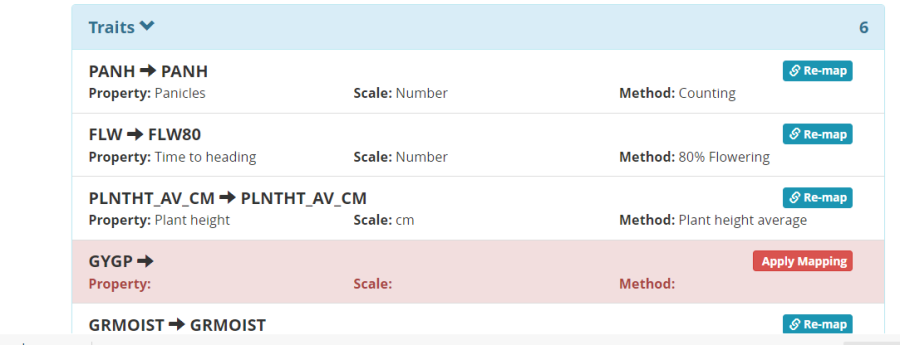
Click Continue to read the layout file and you will open a trait mapping form. Nearly all the traits have been automatically mapped to variables in the ontology because they have exactly the same names. You should check each to be sure the automatic mapping is correct. Any variables which cannot be mapped remail remain in the Unmapped Un-mapped variables box.
In our case GYGP – grain yield in grams per plot has not been mapped. It should be mapped to the variable GrYld_wgh_gplot. To make this mapping drag the variable down and place it in the Traits section then click Apply Mapping.
This will open the Ontology search box. Look for grain yield traits and select GrYld_wgh_gplot.
Now the mapping is complete click Next.
You will get a review panel to check the import:
Click Finish and the design (and data in our case) will be stored.
Add trait GYTHA if necessary and compute its values for all locations.